# Loading our dataset
try:
from palmerpenguins import load_penguins
except:
! pip install palmerpenguins
from palmerpenguins import load_penguins
penguins = load_penguins()
# Pairplot using seaborn
import matplotlib.pyplot as plt
import seaborn as sns
sns.pairplot(penguins, hue='species', markers=["o", "s", "D"])
plt.suptitle("Pairwise Scatter Plots of Penguins Features")
plt.show()Learning Algorithms
CSI 4106 - Fall 2025
Version: Sep 23, 2025 13:43
Preamble
Message of the Day
Learning outcomes
- Differentiate between model, objective, and optimizer in learning algorithms.
- Explain KNN for classification and regression, including uniform and distance-weighted prediction.
- Describe decision trees and apply the split criterion using impurity measures such as Gini.
- Interpret decision boundaries and the concept of linear separability.
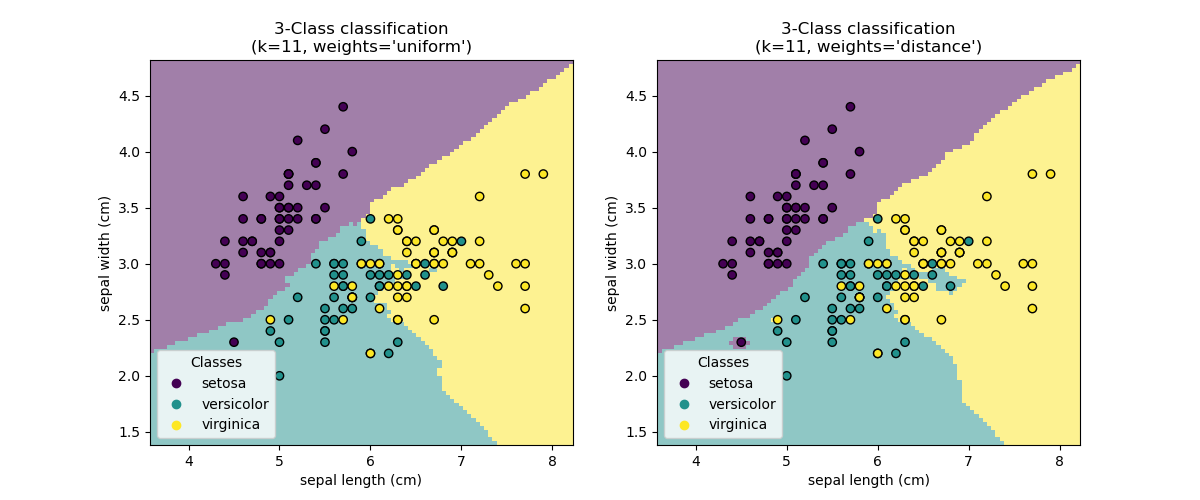
KNN
k-nearest neighbours (KNN)
KNN - Learning
- Lazy learning: no explicit training
- The “model” is the dataset
KNN - Inference
- Classify/regress based on the labels/values of the \(k\) closest examples in feature space
Formal Definition
Given dataset \(\{(x_i, y_i)\}_1^{N}\) and an unseen example \(x\):
- Compute distances \(d(x, x_i)\)
- Select the \(k\) smallest distances
- Classification: majority vote (possibly weighted by \(1/d\))
- Regression: average (possibly weighted)
Exercises
Download these examples to experiment with code variations. Notably, examine how changes in \(k\) impact classification decision boundaries and regression line smoothness.
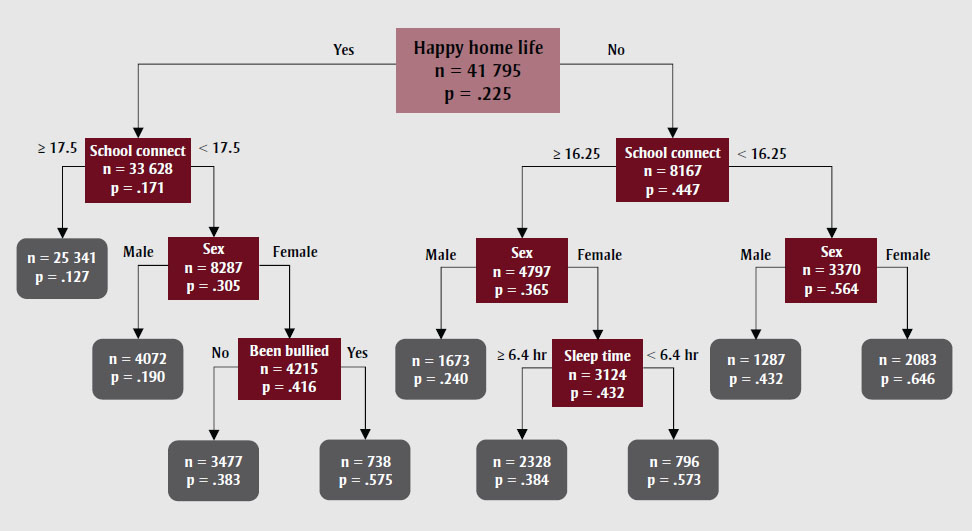
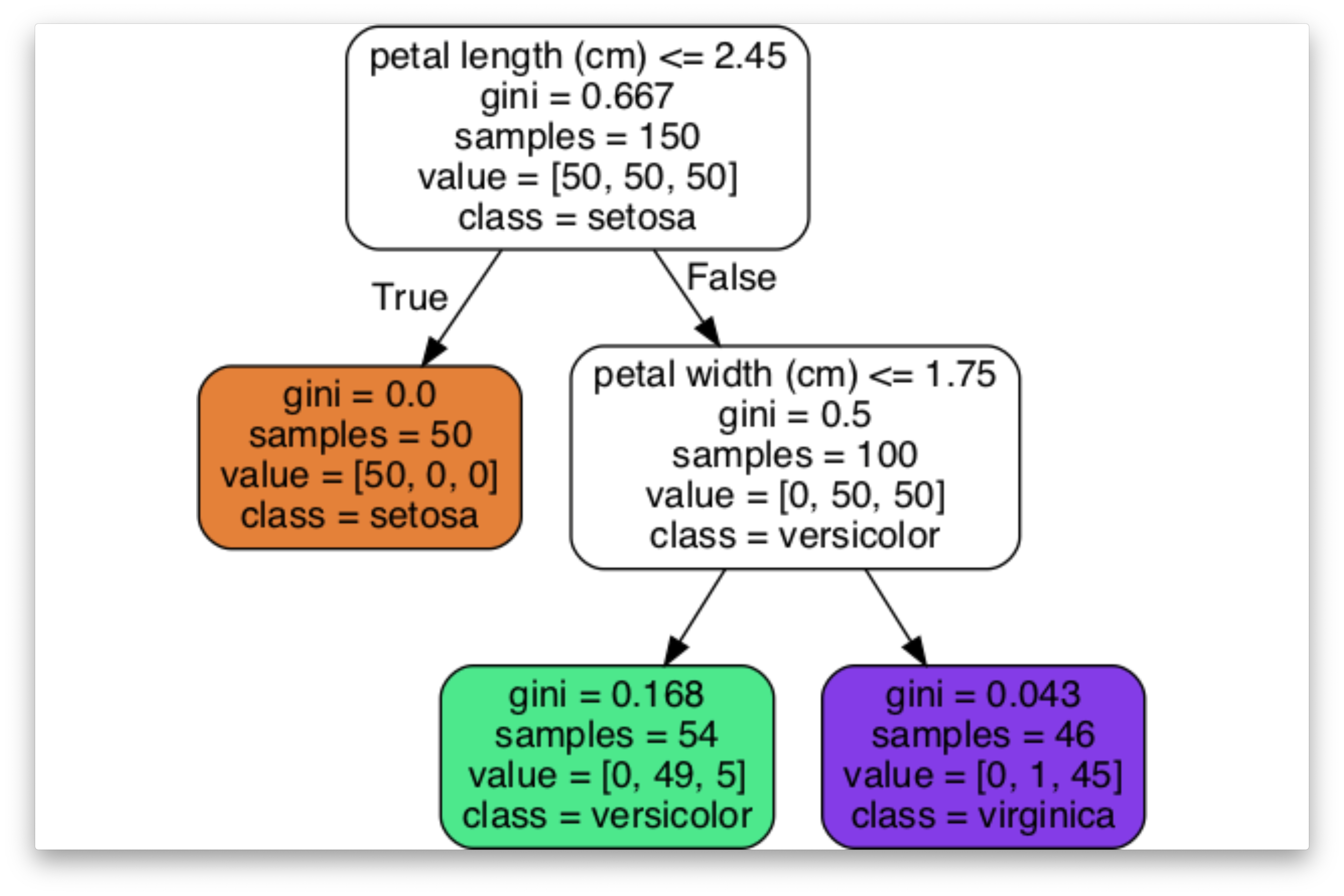
Decision Tree
Interpretable
What is a Decision Tree?
- A decision tree is a hierarchical structure represented as a directed acyclic graph, used for classification and regression tasks.
- Each internal node performs a binary test on a particular feature (\(j\)), such as evaluating whether the number of connections at a school surpasses a specified threshold.
- The leaves function as decision nodes.
Classifying New Instances (Inference)
- Begin at the root node of the decision tree. Proceed by answering a sequence of binary questions until a leaf node is reached. The label associated with this leaf denotes the classification of the instance.
- Alternatively, some algorithms may store a probability distribution at the leaf, representing the fraction of training samples corresponding to each class \(k\), across all possible classes \(k\).
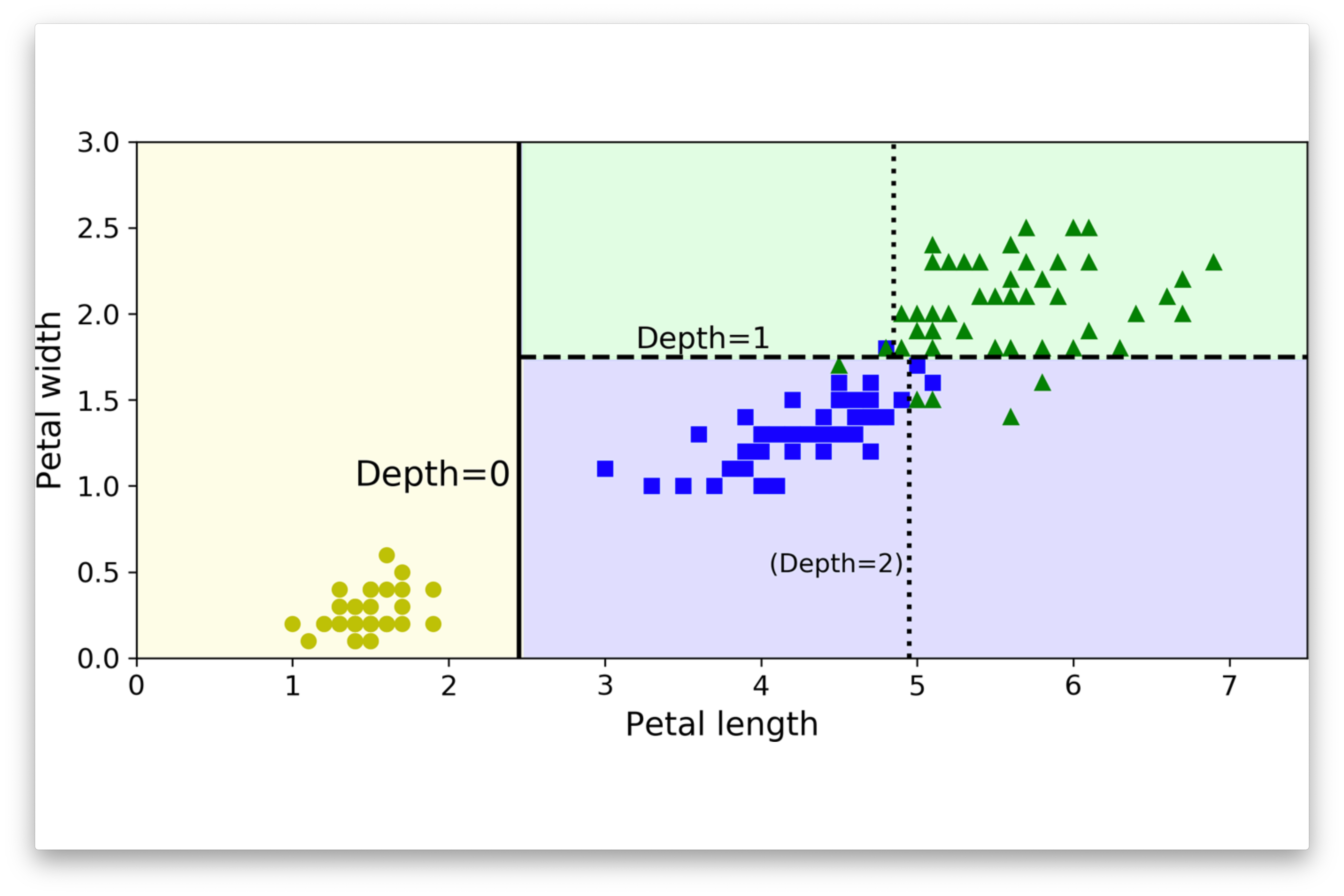
Decision Boundary
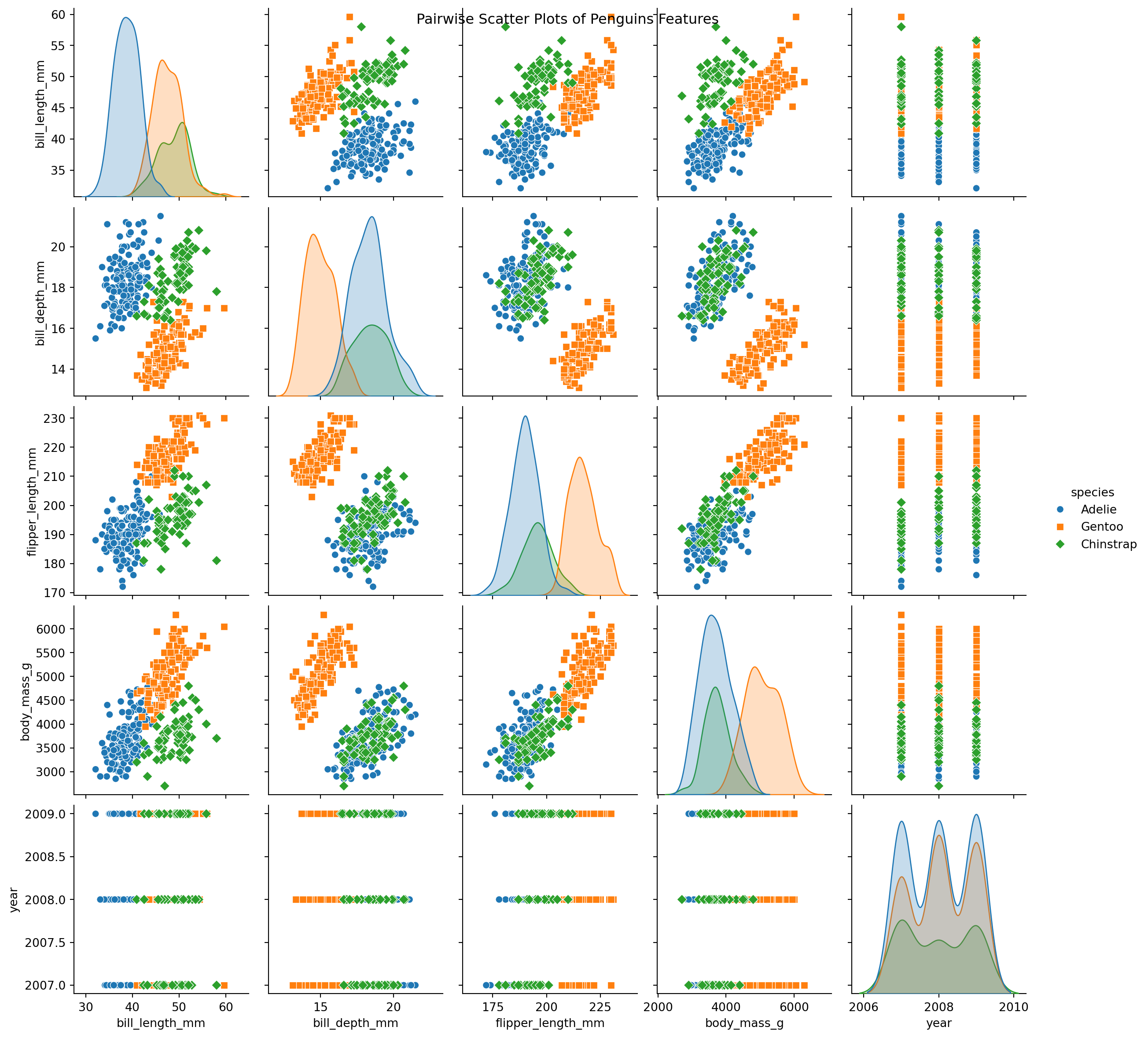
Palmer Pinguins Dataset
Palmer Pinguins Dataset
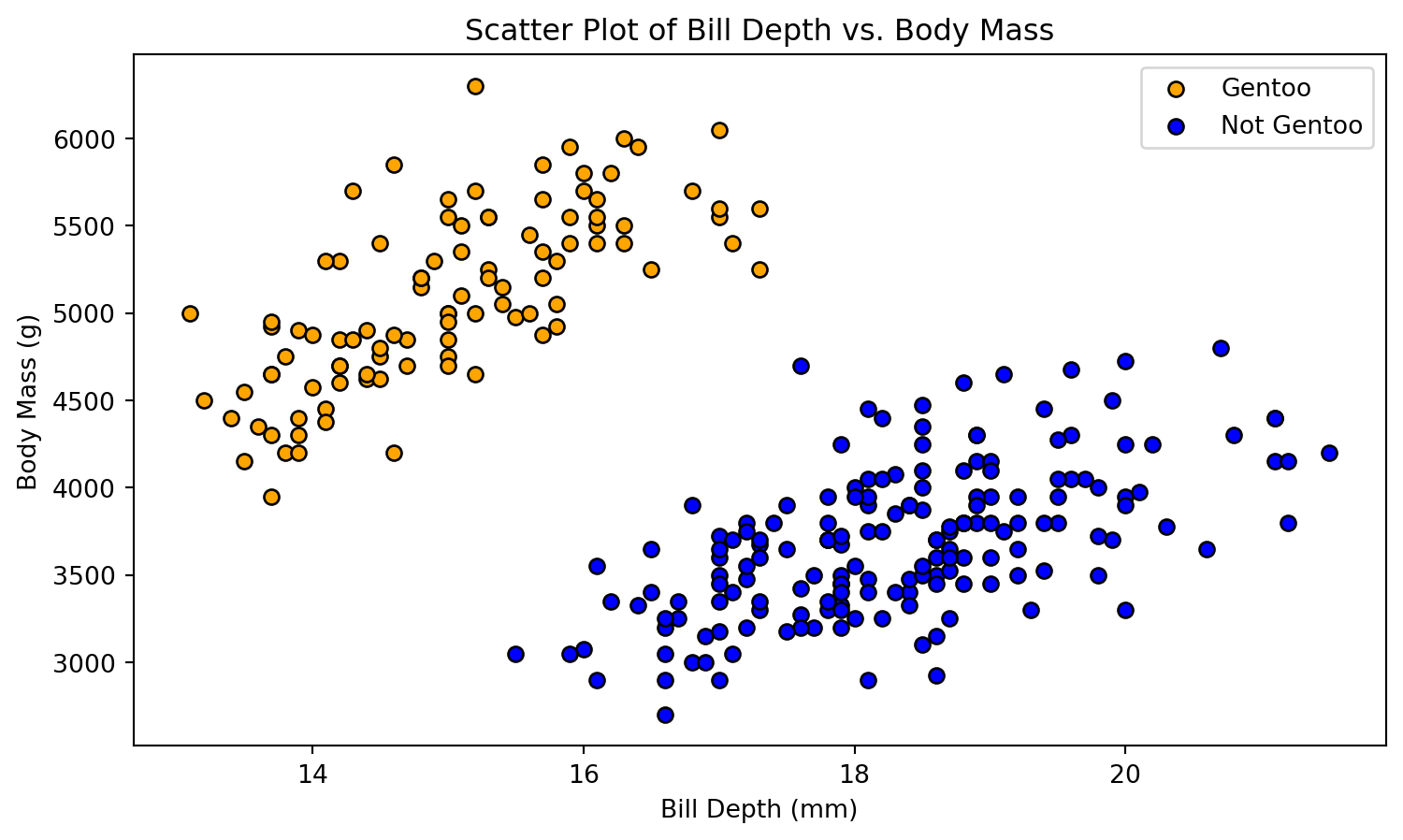
Binary Classification Problem
- Several scatter plots reveal a distinct clustering of Gentoo instances.
- To illustrate our next exemple, we propose a binary classification model: Gentoo versus non-Gentoo.
- Our analysis will concentrate on two key features: body mass and bill depth.
Definition
A decision boundary is a “boundary” that partitions the underlying feature space into regions corresponding to different class labels.
Decision Boundary
The decision boundary between these attributes can be represented as a line.
Code
# Import necessary libraries
import numpy as np
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
try:
from palmerpenguins import load_penguins
except:
! pip install palmerpenguins
from palmerpenguins import load_penguins
# Load the Palmer Penguins dataset
df = load_penguins()
# Preserve only the necessary features: 'bill_depth_mm' and 'body_mass_g'
features = ['bill_depth_mm', 'body_mass_g']
df = df[features + ['species']]
# Drop rows with missing values
df.dropna(inplace=True)
# Create a binary problem: 'Gentoo' vs 'Not Gentoo'
df['species_binary'] = df['species'].apply(lambda x: 1 if x == 'Gentoo' else 0)
# Define feature matrix X and target vector y
X = df[features].values
y = df['species_binary'].values
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Function to plot initial scatter of data
def plot_scatter(X, y):
plt.figure(figsize=(9, 5))
plt.scatter(X[y == 1, 0], X[y == 1, 1], color='orange', edgecolors='k', marker='o', label='Gentoo')
plt.scatter(X[y == 0, 0], X[y == 0, 1], color='blue', edgecolors='k', marker='o', label='Not Gentoo')
plt.xlabel('Bill Depth (mm)')
plt.ylabel('Body Mass (g)')
plt.title('Scatter Plot of Bill Depth vs. Body Mass')
plt.legend()
plt.show()
# Plot the initial scatter plot
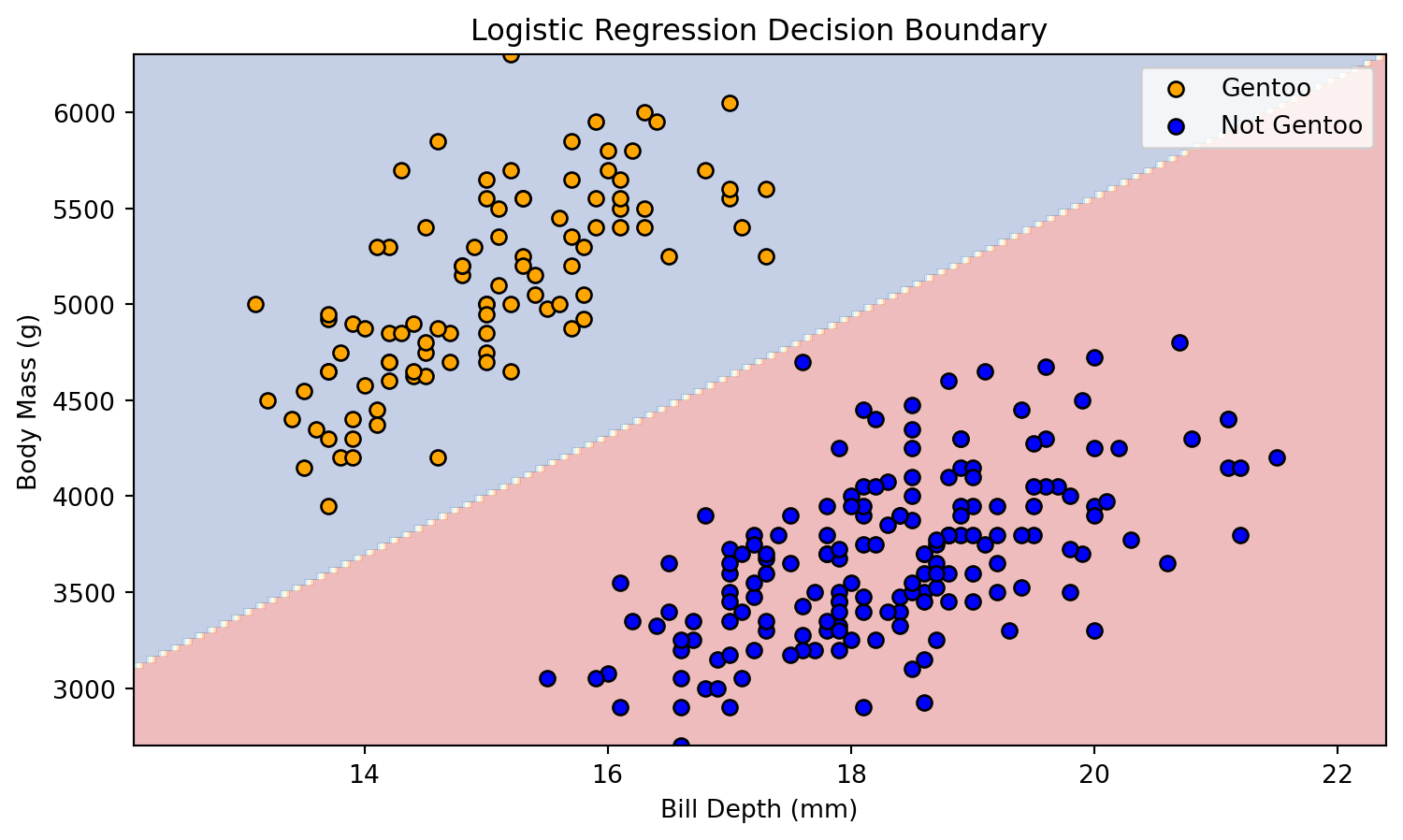
plot_scatter(X_train, y_train)Decision Boundary
Decision Boundary
The decision boundary between these attributes can be represented as a line.
Code
# Train a logistic regression model
model = LogisticRegression()
model.fit(X_train, y_train)
# Function to plot decision boundary
def plot_decision_boundary(X, y, model):
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.1),
np.arange(y_min, y_max, 0.1))
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.figure(figsize=(9, 5))
plt.contourf(xx, yy, Z, alpha=0.3, cmap='RdYlBu')
plt.scatter(X[y == 1, 0], X[y == 1, 1], color='orange', edgecolors='k', marker='o', label='Gentoo')
plt.scatter(X[y == 0, 0], X[y == 0, 1], color='blue', edgecolors='k', marker='o', label='Not Gentoo')
plt.xlabel('Bill Depth (mm)')
plt.ylabel('Body Mass (g)')
plt.title('Logistic Regression Decision Boundary')
plt.legend()
plt.show()
# Plot the decision boundary on the training set
plot_decision_boundary(X_train, y_train, model)Decision Boundary
Definition
We say that the data is linearly separable when two classes of data can be perfectly separated by a single linear boundary, such as a line in two-dimensional space or a hyperplane in higher dimensions.
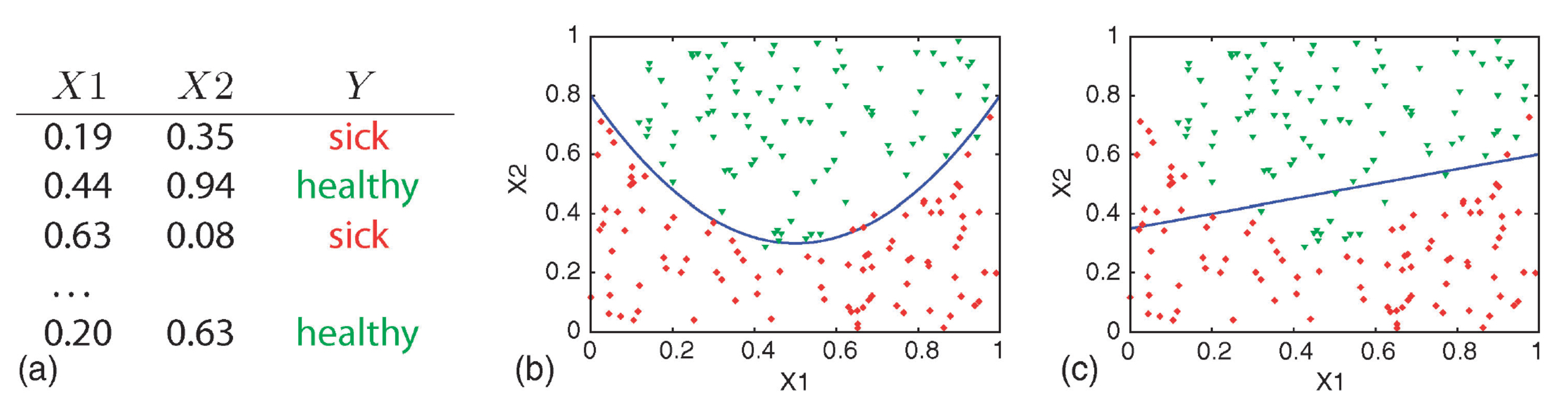
Simple Decision Doundary
(a) training data, (b) quadratic curve, and (c) linear function.
Attribution: (Geurts, Irrthum, and Wehenkel 2009)
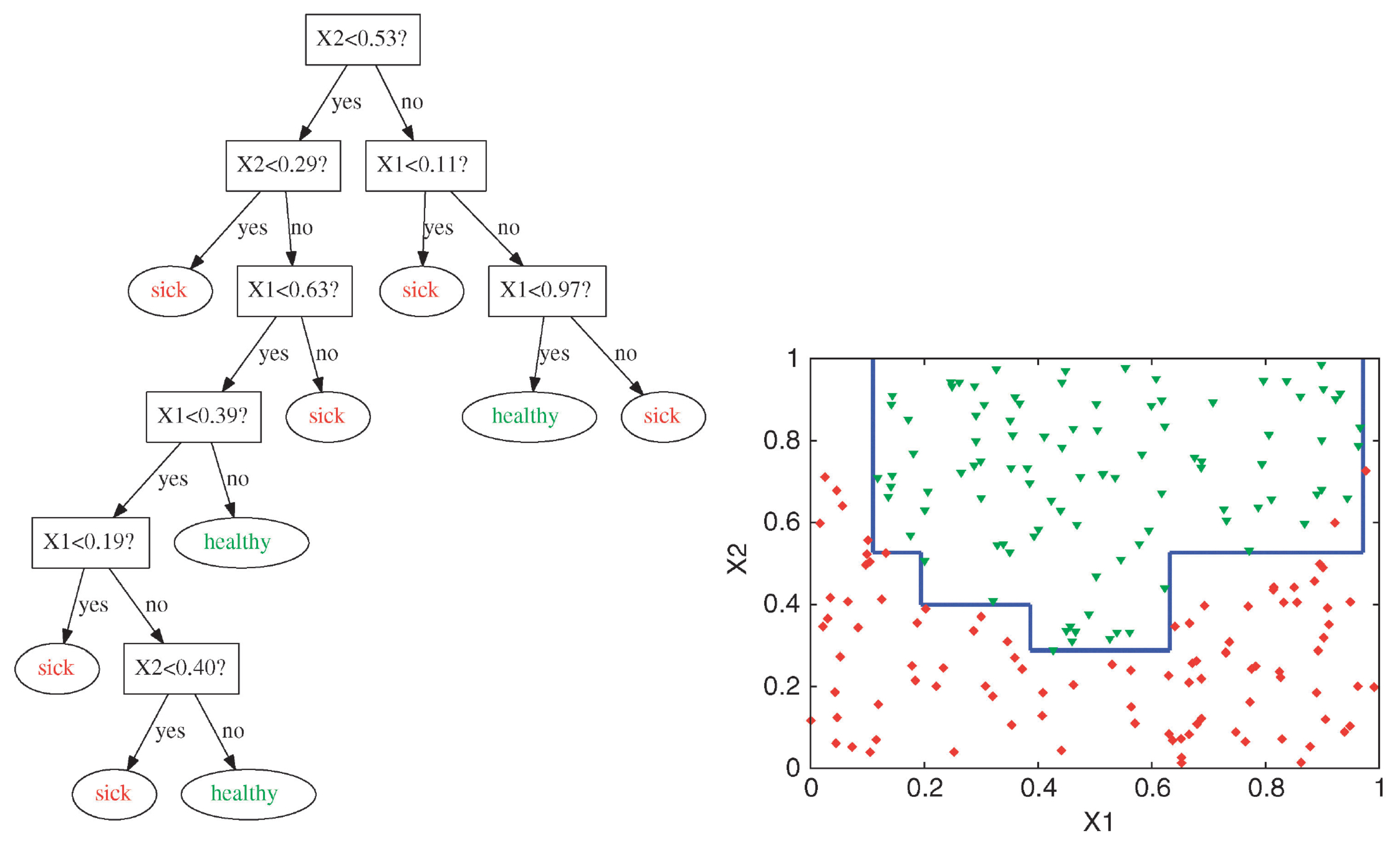
Complex Decision Boundary
Decision trees are capable of generating irregular and non-linear decision boundaries.
Attribution: ibidem.
Definition (revised)
A decision boundary is a hypersurface that partitions the underlying feature space into regions corresponding to different class labels.
Decision Tree (contd)
Constructing a Decision Tree
How to construct (learnt) a decision tree?
Are there some trees that are “better” than others?
Is it feasible to construct an optimal decision tree with computational efficiency?
Optimality
- Let \(X = \{x_1, \ldots, x_n\}\) be a finite set of objects.
- Let \(\mathcal{T} = \{T_1, \ldots, T_t\}\) be a finite set of tests.
- For each object and test, we have:
- \(T_i(x_j)\) is either true or false.
- An optimal tree is one that completely identifies all the objects in \(X\) and \(|T|\) is minimum.
Constructing a Decision Tree
- Iterative development: Initiate with an empty tree. Progressively introduce nodes, each informed by the training dataset, continuing until the dataset is completely classified or alternative termination criteria, such as maximum tree depth, are met.
Constructing a Decision Tree
- Initial Node Construction:
- To establish the root node, evaluate all available \(D\) features.
- For each feature, assess various threshold values derived from the observed data within the training set.
- To establish the root node, evaluate all available \(D\) features.
Constructing a Decision Tree
- For a numerical feature, the algorithm considers all possible split points (thresholds) in the feature’s range.
- These split points are typically the midpoints between two consecutive, sorted unique values of the feature.
Constructing a Decision Tree
- For a categorical feature with \(k\) unique values, the algorithm considers all possible ways of splitting the categories into two groups.
- For instance, if the feature (forecast) has values, ‘Rainy’, ‘Cloudy’, and ‘Sunny’, it evaluates the following splits:
- \(\{\mathrm{Rainy}\}\) vs. \(\{\mathrm{Cloudy}, \mathrm{Sunny}\}\),
- \(\{\mathrm{Cloudy}\}\) vs. \(\{\mathrm{Rainy}, \mathrm{Sunny}\}\) ,
- \(\{\mathrm{Sunny}\}\) vs. \(\{\mathrm{Rainy}, \mathrm{Cloudy}\}\).
Evaluation
What defines a “good” data split?
- \(\{\mathrm{Rainy}\}\) vs. \(\{\mathrm{Cloudy}, \mathrm{Sunny}\}\) : \([20,10,5]\) and \([10,10,15]\).
- \(\{\mathrm{Cloudy}\}\) vs. \(\{\mathrm{Rainy}, \mathrm{Sunny}\}\) : \([40,0,0]\) and \([0,30,0]\).
Evaluation
Heterogeneity (also referred to as impurity) and homogeneity are critical metrics for evaluating the composition of resulting data partitions.
Optimally, each of these partitions should contain data entries from a single class to achieve maximum homogeneity.
Entropy and the Gini index are two widely utilized metrics for assessing these characteristics.
Evalution
Objective function for sklearn.tree.DecisionTreeClassifier (CART):
\[ J(k,t_k) = \frac{N_{\text{left}}}{N_{\text{parent}}} G_{\text{left}} + \frac{N_{\text{right}}}{N_{\text{parent}}} G_{\text{right}} \]
The cost of partitioning the data using feature \(k\) and threshold \(t_k\).
\(N_{\text{left}}\) and \(N_{\text{right}}\) is the number of examples in the left and right subsets, respectively, and \(N_{\text{parent}}\) is the number of examples before splitting the data.
\(G_{\text{left}}\) and \(G_{\text{right}}\) is the impurity of the left and right subsets, respectively.
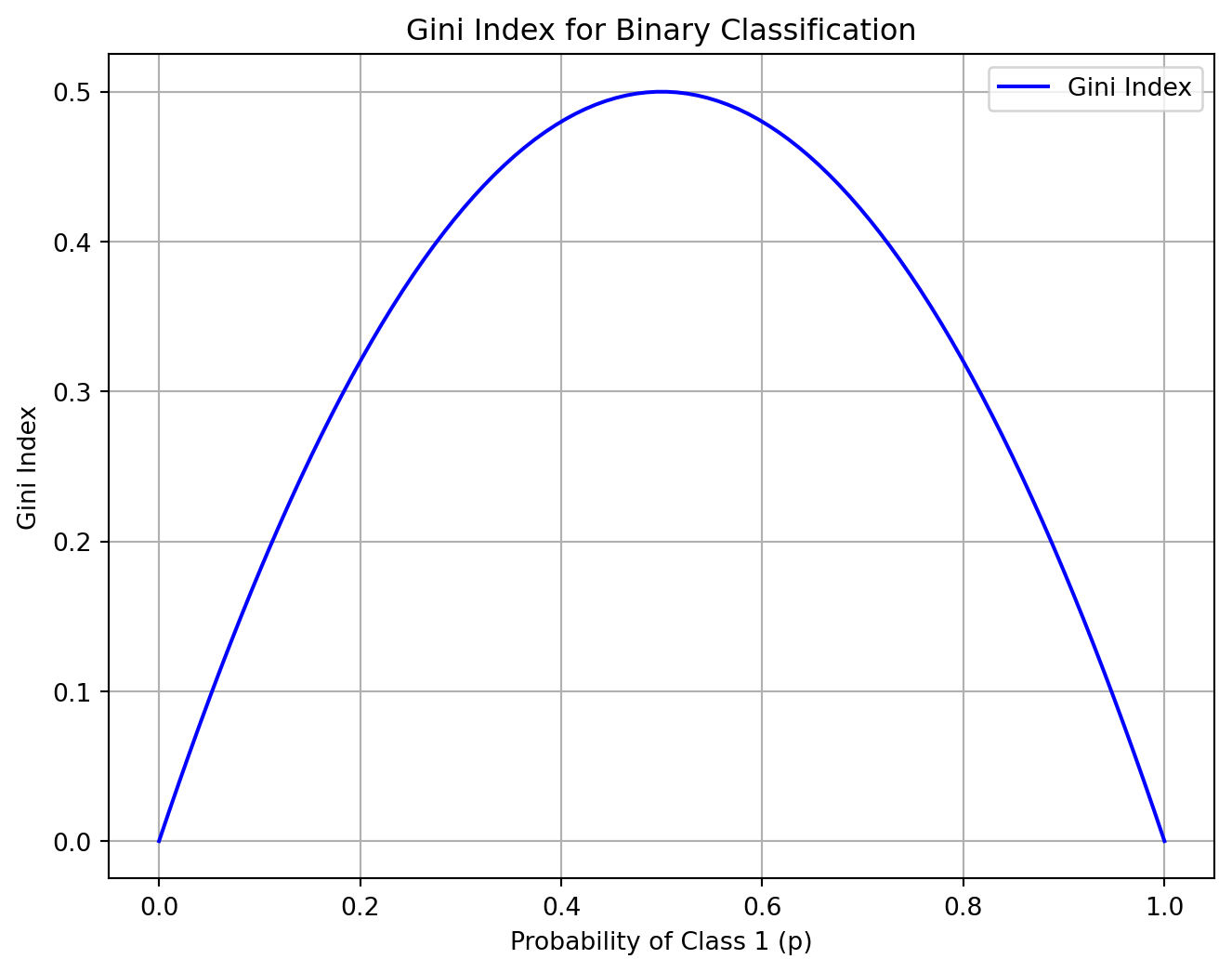
Gini Index
- Gini index (default)
\[ G_i = 1 - \sum_{k=1}^n p_{i,k}^2 \]
\(p_{i,k}\) is the proportion of the examples from this class \(k\) in the node \(i\).
What is the maximum value of the Gini index?
Gini Index
Considering a binary classification problem:
- \(1 - [(0/100)^2 + (100/100)^2] = 0\) (pure)
- \(1 - [(25/100)^2 + (75/100)^2] = 0.375\)
- \(1 - [(50/100)^2 + (50/100)^2] = 0.5\)
Gini Index
Code
def gini_index(p):
"""Calculate the Gini index."""
return 1 - (p**2 + (1 - p)**2)
# Probability values for class 1
p_values = np.linspace(0, 1, 100)
# Calculate Gini index for each probability
gini_values = [gini_index(p) for p in p_values]
# Plot the Gini index
plt.figure(figsize=(8, 6))
plt.plot(p_values, gini_values, label='Gini Index', color='b')
plt.title('Gini Index for Binary Classification')
plt.xlabel('Probability of Class 1 (p)')
plt.ylabel('Gini Index')
plt.grid(True)
plt.legend()
plt.show()Iris Dataset
Complete Example
Stopping Criteria
- All the examples in a given node belong to the same class.
- Depth of the tree would exceed max_depth.
- Number of examples in the node is min_sample_split or less.
- None of the splits decreases impurity sufficiently (min_impurity_decrease).
- See documentation for other criteria.
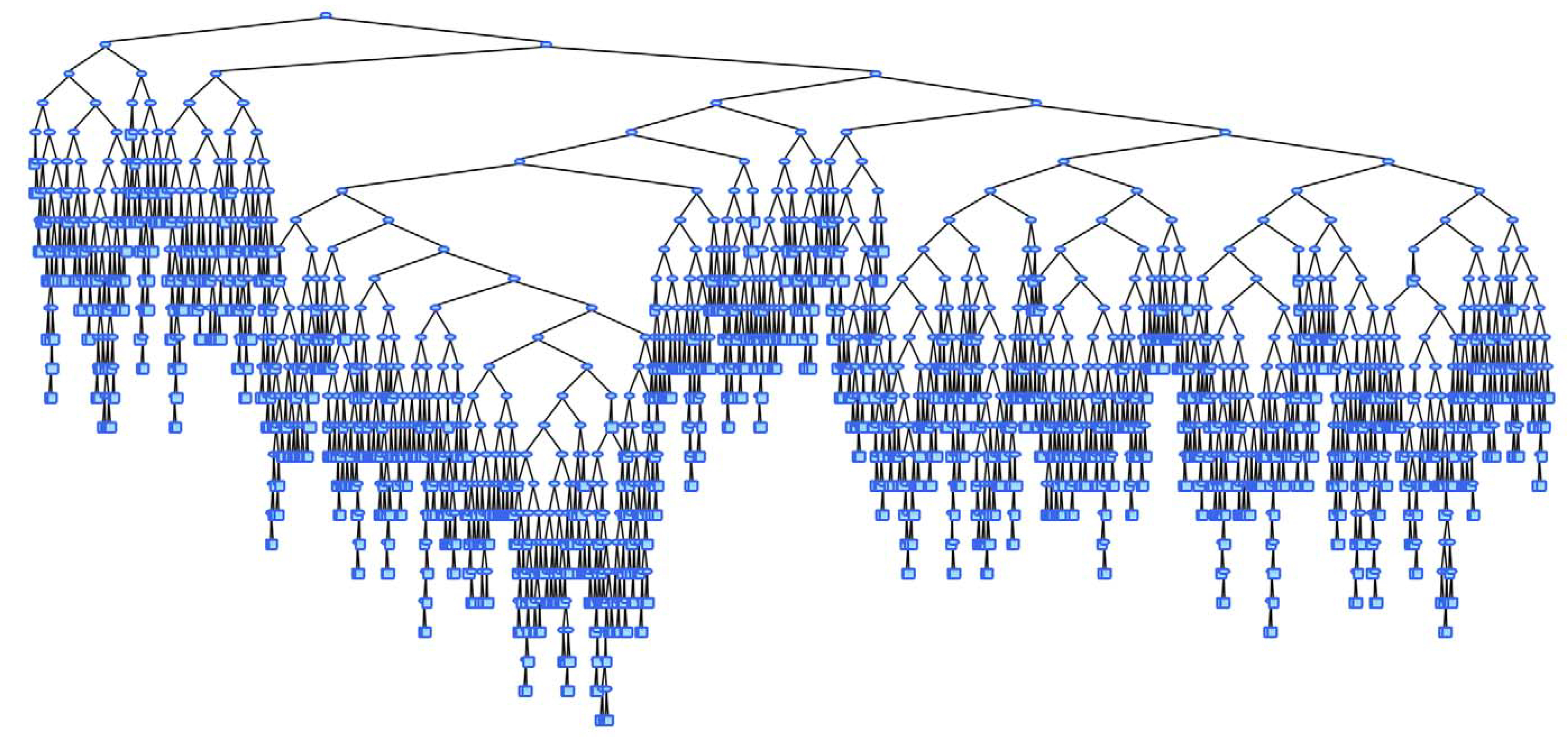
Limitations
- Possibly creates large trees
- Challenge for interpretation
- Overfitting
- Greedy algorithm, no guarantee to find the optimal tree. (Hyafil and Rivest 1976)
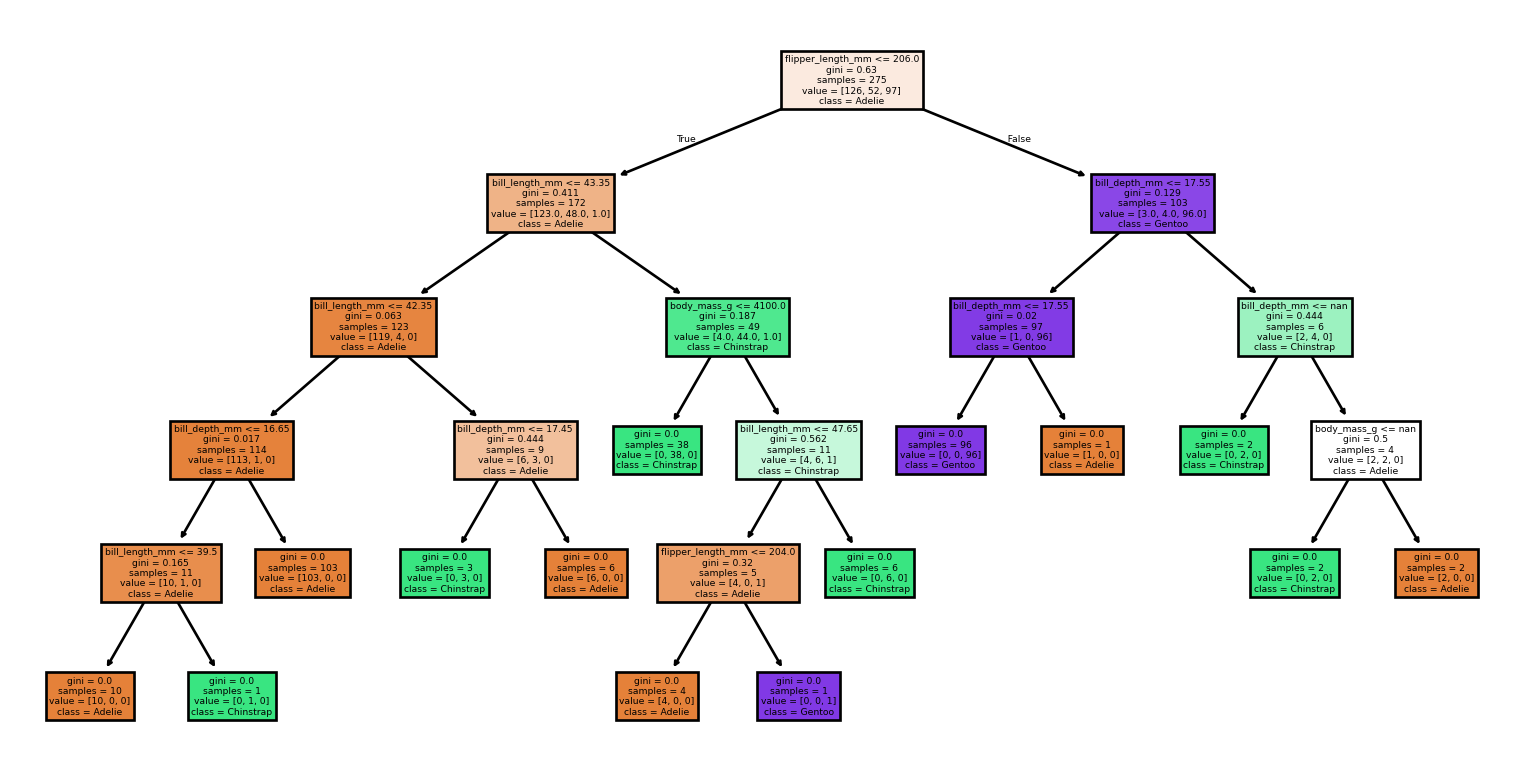
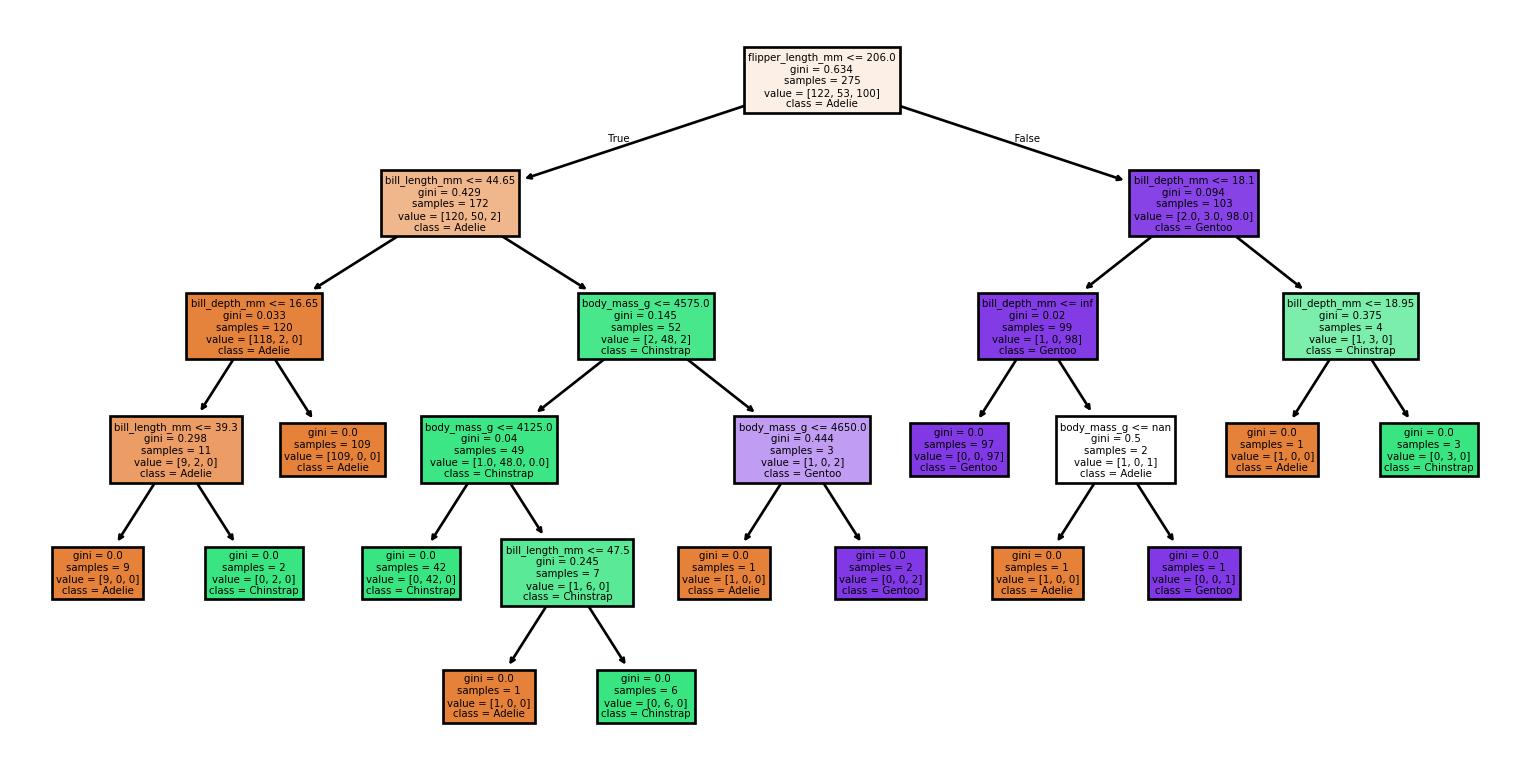
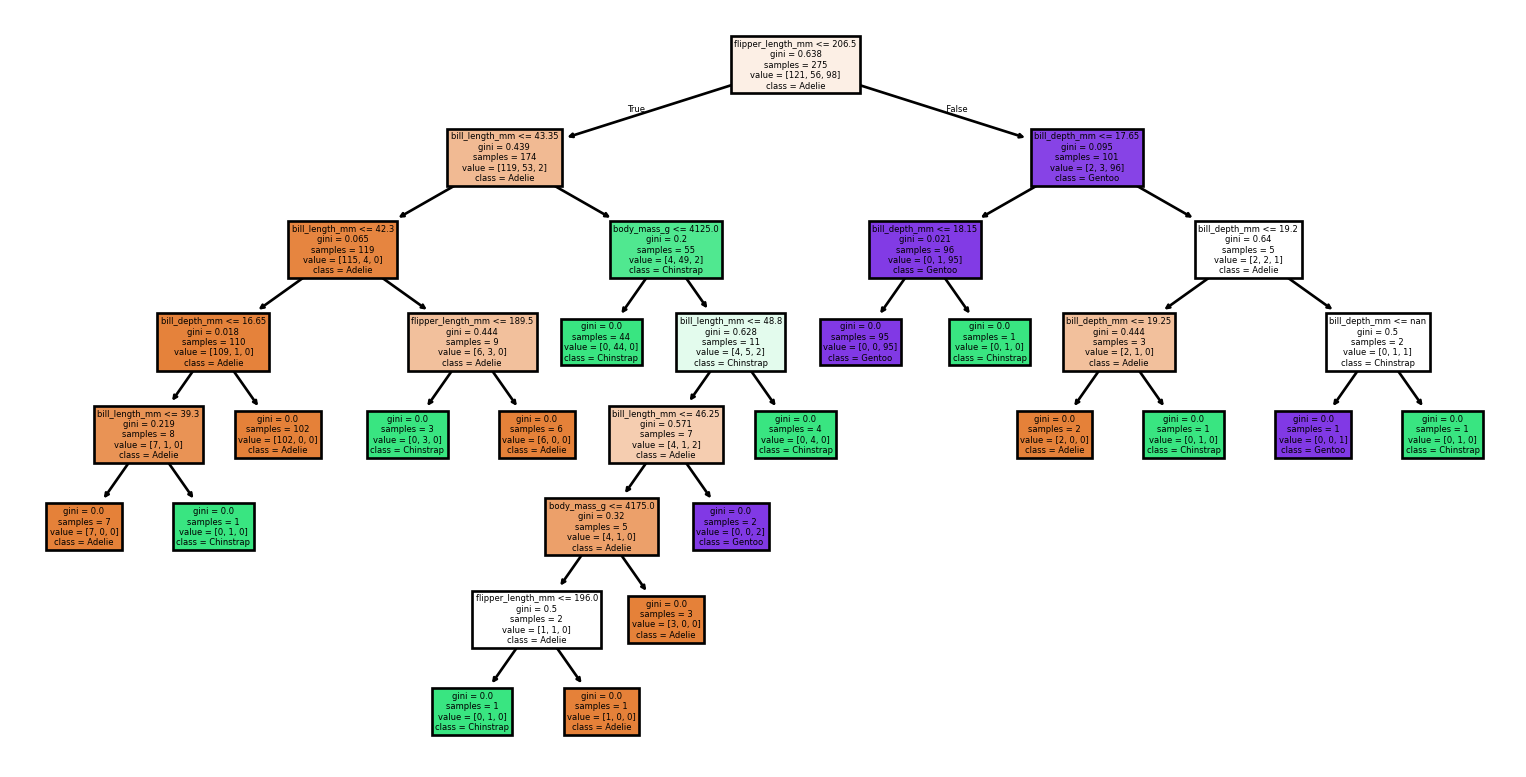
- Small changes to the data set produce vastly different trees
Large Trees
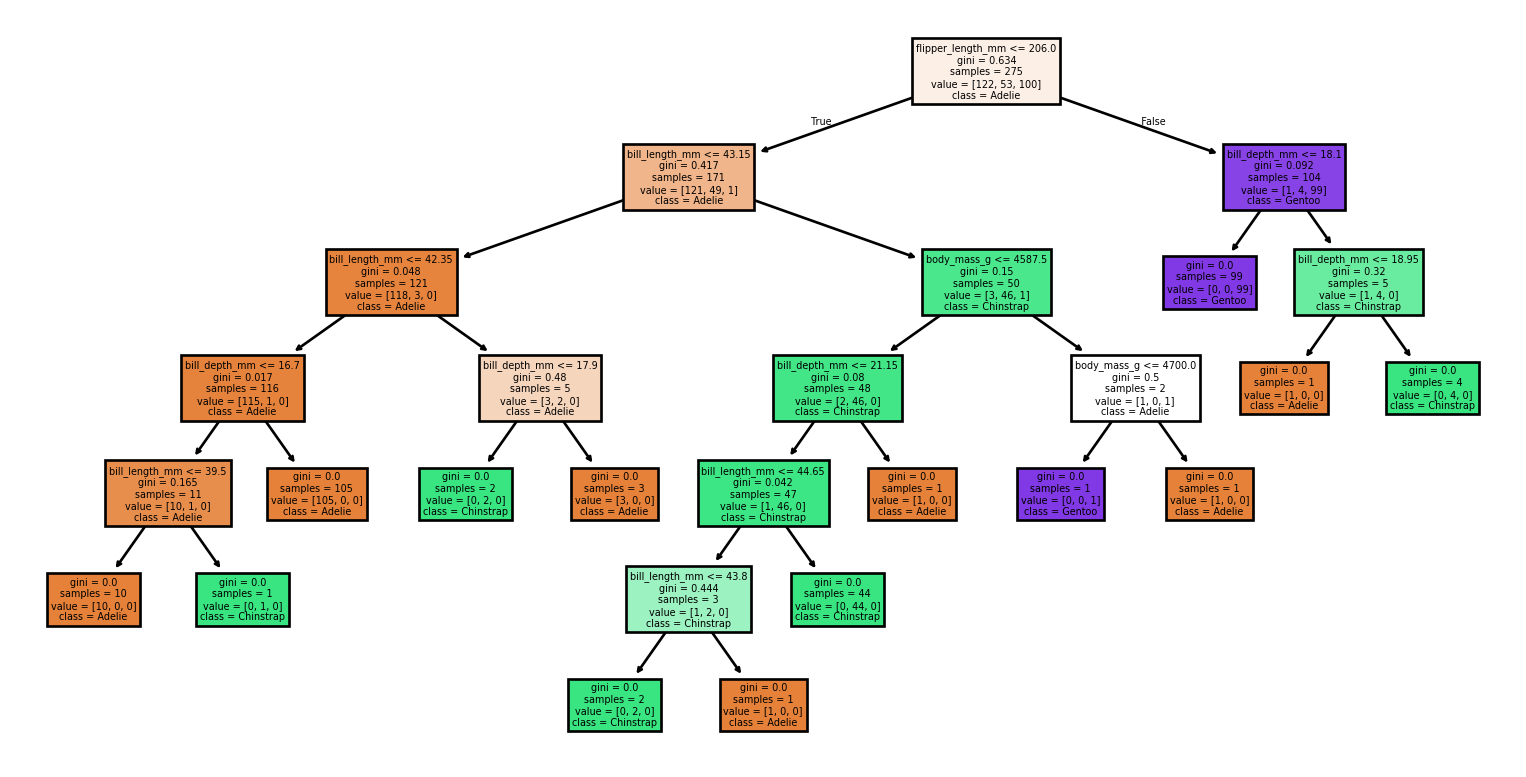
Small Changes to the Dataset
Code
from sklearn import tree
from sklearn.metrics import classification_report, accuracy_score
# Loading the dataset
X, y = load_penguins(return_X_y = True)
target_names = ['Adelie','Chinstrap','Gentoo']
# Split the dataset into training and testing sets
for seed in (4, 7, 90, 96, 99, 2):
print(f'Seed: {seed}')
# Create new training and test sets based on a different random seed
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=seed)
# Creating a new classifier
clf = tree.DecisionTreeClassifier(random_state=seed)
# Training
clf.fit(X_train, y_train)
# Make predictions
y_pred = clf.predict(X_test)
# Plotting the tree
tree.plot_tree(clf,
feature_names = X.columns,
class_names = target_names,
filled = True)
plt.show()
# Evaluating the model
accuracy = accuracy_score(y_test, y_pred)
report = classification_report(y_test, y_pred, target_names=target_names)
print(f'Accuracy: {accuracy:.2f}')
print('Classification Report:')
print(report)Small Changes to the Dataset
Seed: 4Accuracy: 0.99
Classification Report:
precision recall f1-score support
Adelie 1.00 0.97 0.99 36
Chinstrap 0.94 1.00 0.97 17
Gentoo 1.00 1.00 1.00 16
accuracy 0.99 69
macro avg 0.98 0.99 0.99 69
weighted avg 0.99 0.99 0.99 69
Seed: 7Accuracy: 0.91
Classification Report:
precision recall f1-score support
Adelie 0.96 0.83 0.89 30
Chinstrap 0.83 1.00 0.91 15
Gentoo 0.92 0.96 0.94 24
accuracy 0.91 69
macro avg 0.90 0.93 0.91 69
weighted avg 0.92 0.91 0.91 69
Seed: 90Accuracy: 0.94
Classification Report:
precision recall f1-score support
Adelie 0.90 1.00 0.95 26
Chinstrap 0.93 0.88 0.90 16
Gentoo 1.00 0.93 0.96 27
accuracy 0.94 69
macro avg 0.94 0.93 0.94 69
weighted avg 0.95 0.94 0.94 69
Seed: 96Accuracy: 0.90
Classification Report:
precision recall f1-score support
Adelie 0.83 0.97 0.89 30
Chinstrap 1.00 0.67 0.80 15
Gentoo 0.96 0.96 0.96 24
accuracy 0.90 69
macro avg 0.93 0.86 0.88 69
weighted avg 0.91 0.90 0.90 69
Seed: 99Accuracy: 1.00
Classification Report:
precision recall f1-score support
Adelie 1.00 1.00 1.00 31
Chinstrap 1.00 1.00 1.00 12
Gentoo 1.00 1.00 1.00 26
accuracy 1.00 69
macro avg 1.00 1.00 1.00 69
weighted avg 1.00 1.00 1.00 69
Seed: 2Accuracy: 0.55
Classification Report:
precision recall f1-score support
Adelie 0.62 0.97 0.75 30
Chinstrap 0.43 0.90 0.58 10
Gentoo 0.00 0.00 0.00 29
accuracy 0.55 69
macro avg 0.35 0.62 0.44 69
weighted avg 0.33 0.55 0.41 69
Summary
- The lecture surveyed three learning algorithms, k-nearest neighbours (KNN), decision trees, and linear regression, and framed them via model, objective, and optimization.
- We then constructed decision trees, showed that regression leaves returned the sample mean, minimized the weighted impurity \(J\), and analyzed the Gini index.
- Decision boundaries were illustrated for linear and non-linear models.
Prologue
Resources
- Plot the decision surface of decision trees trained on the iris dataset from
sklearn - Decision trees by Jan Kirenz, a Professor at HdM Stuttgart
- CS 320 Apr12-2021 (Part 2) - Decision Boundaries by Tyler Caraza-Harter, an Instructor at UW-Madison
References
Next lecture
- Training a linear model
Marcel Turcotte
School of Electrical Engineering and Computer Science (EECS)
University of Ottawa