Code
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
try:
from palmerpenguins import load_penguins
except ImportError:
! pip install palmerpenguins
from palmerpenguins import load_penguins
# Load the Palmer Penguins dataset
df = load_penguins()
# Keep only 'flipper_length_mm' and 'species'
df = df[['flipper_length_mm', 'species']]
# Drop rows with missing values (NaNs)
df.dropna(inplace=True)
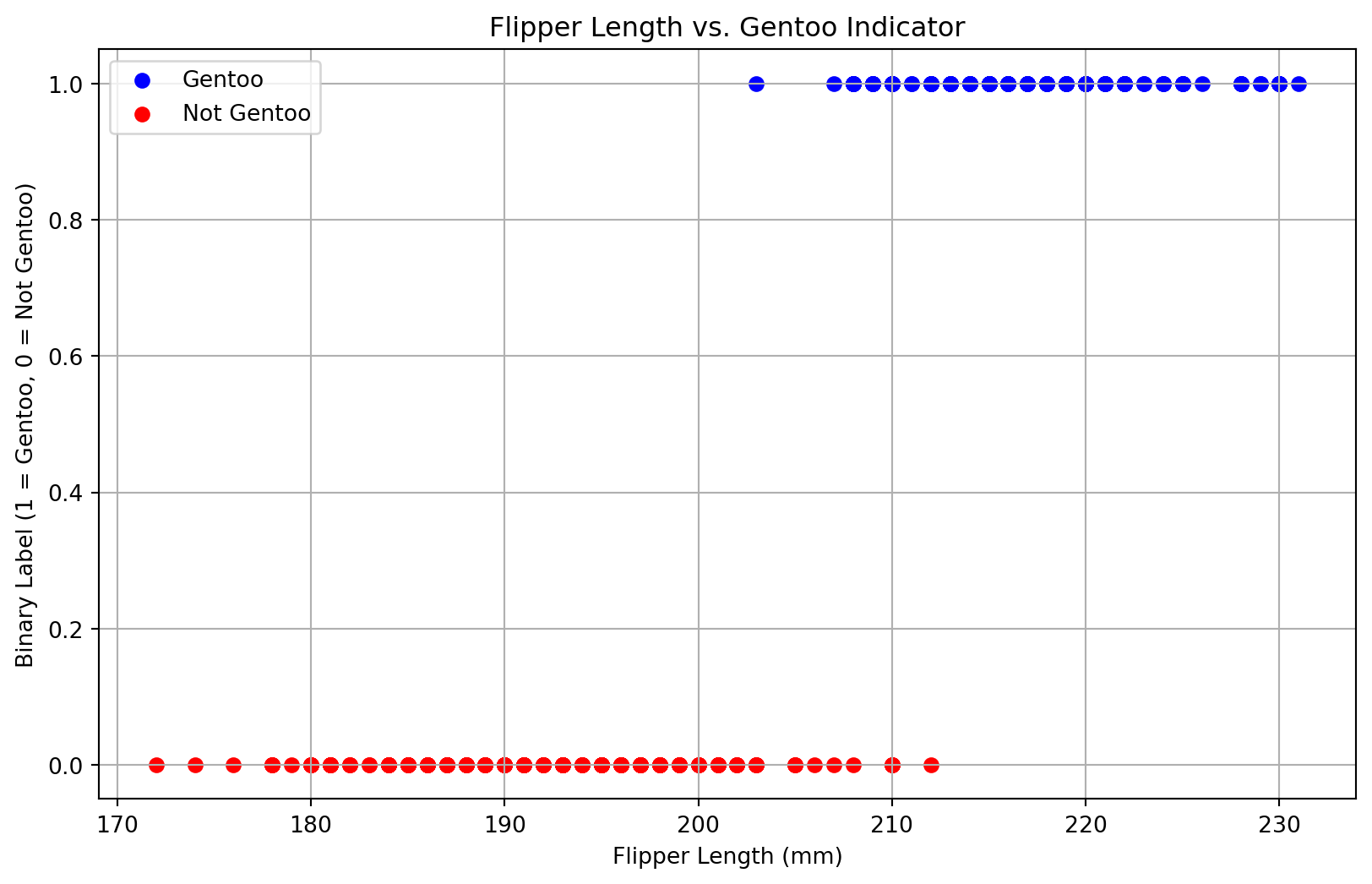
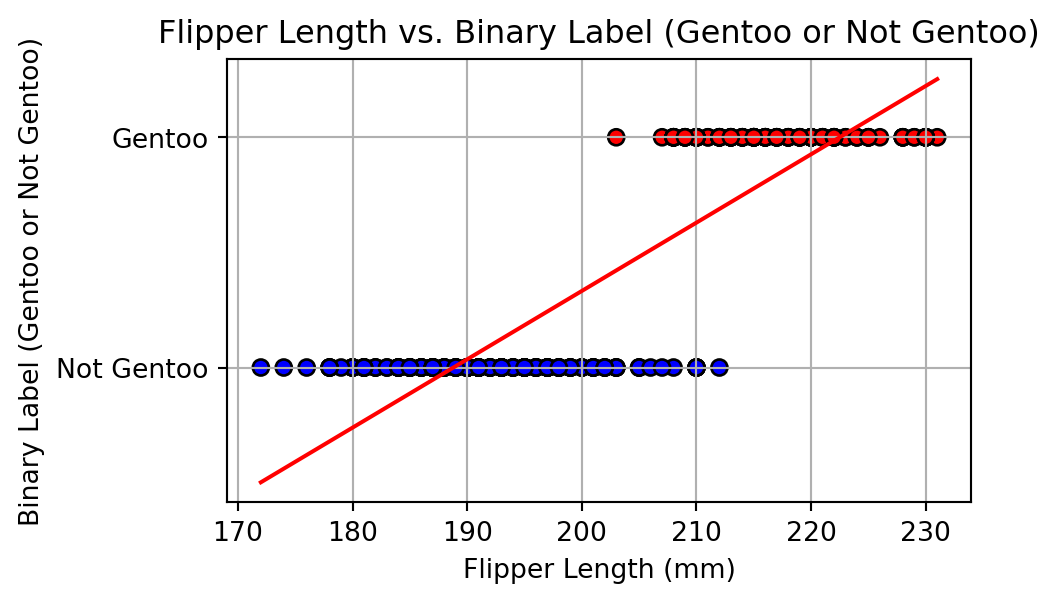
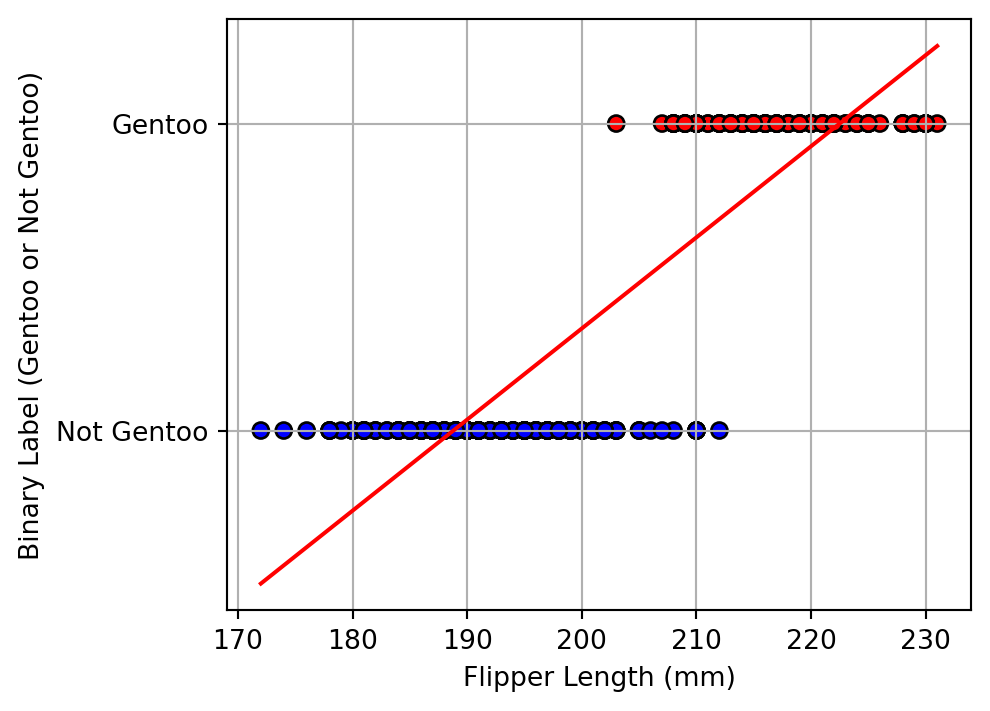
# Create a binary label: 1 if Gentoo, 0 otherwise
df['is_gentoo'] = (df['species'] == 'Gentoo').astype(int)
# Separate features (X) and labels (y)
X = df[['flipper_length_mm']]
y = df['is_gentoo']
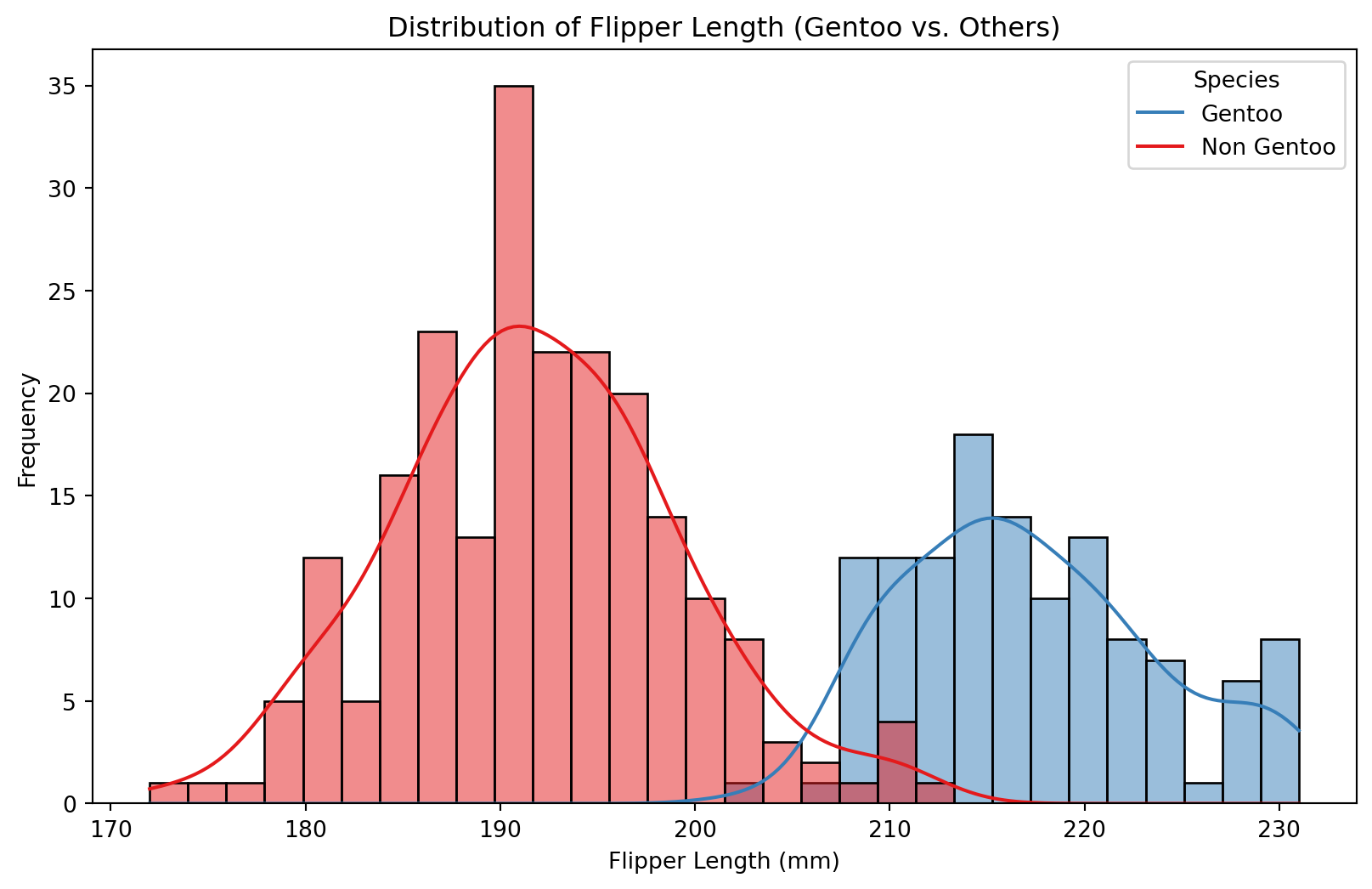
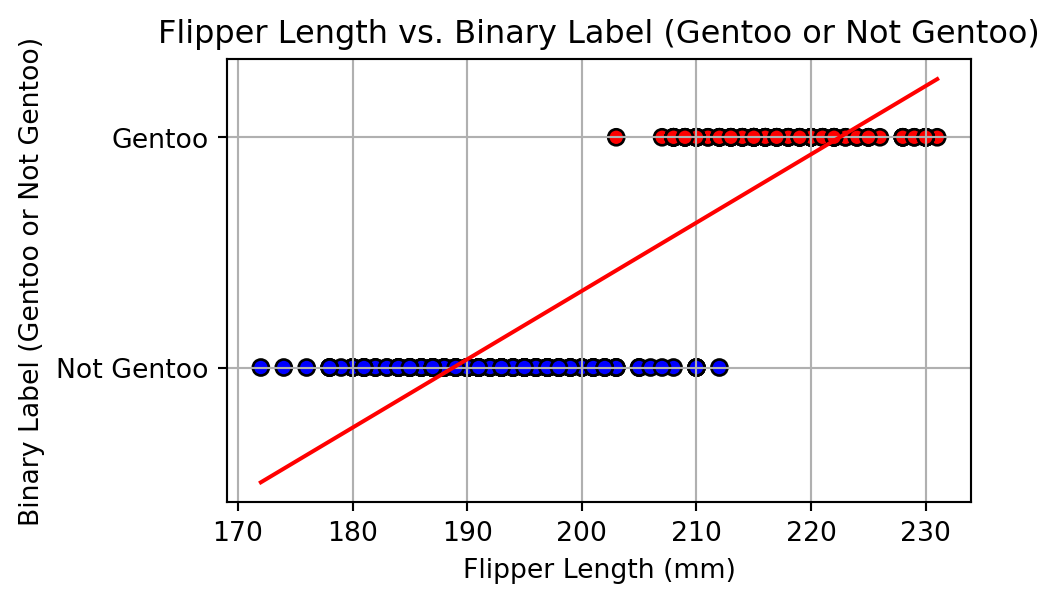
# Plot the distribution of flipper lengths by binary species label
plt.figure(figsize=(10, 6))
sns.histplot(data=df, x='flipper_length_mm', hue='is_gentoo', kde=True, bins=30, palette='Set1')
plt.title('Distribution of Flipper Length (Gentoo vs. Others)')
plt.xlabel('Flipper Length (mm)')
plt.ylabel('Frequency')
plt.legend(title='Species', labels=['Gentoo', 'Non Gentoo'])
plt.show()