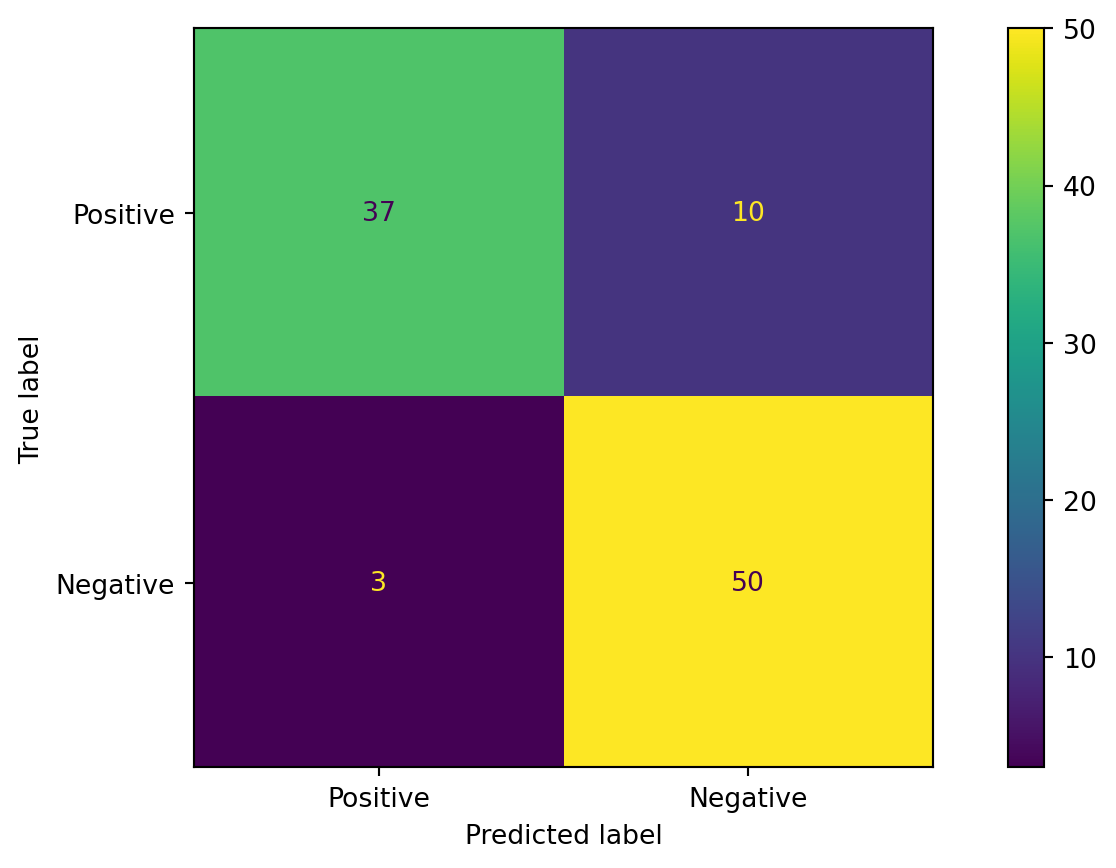
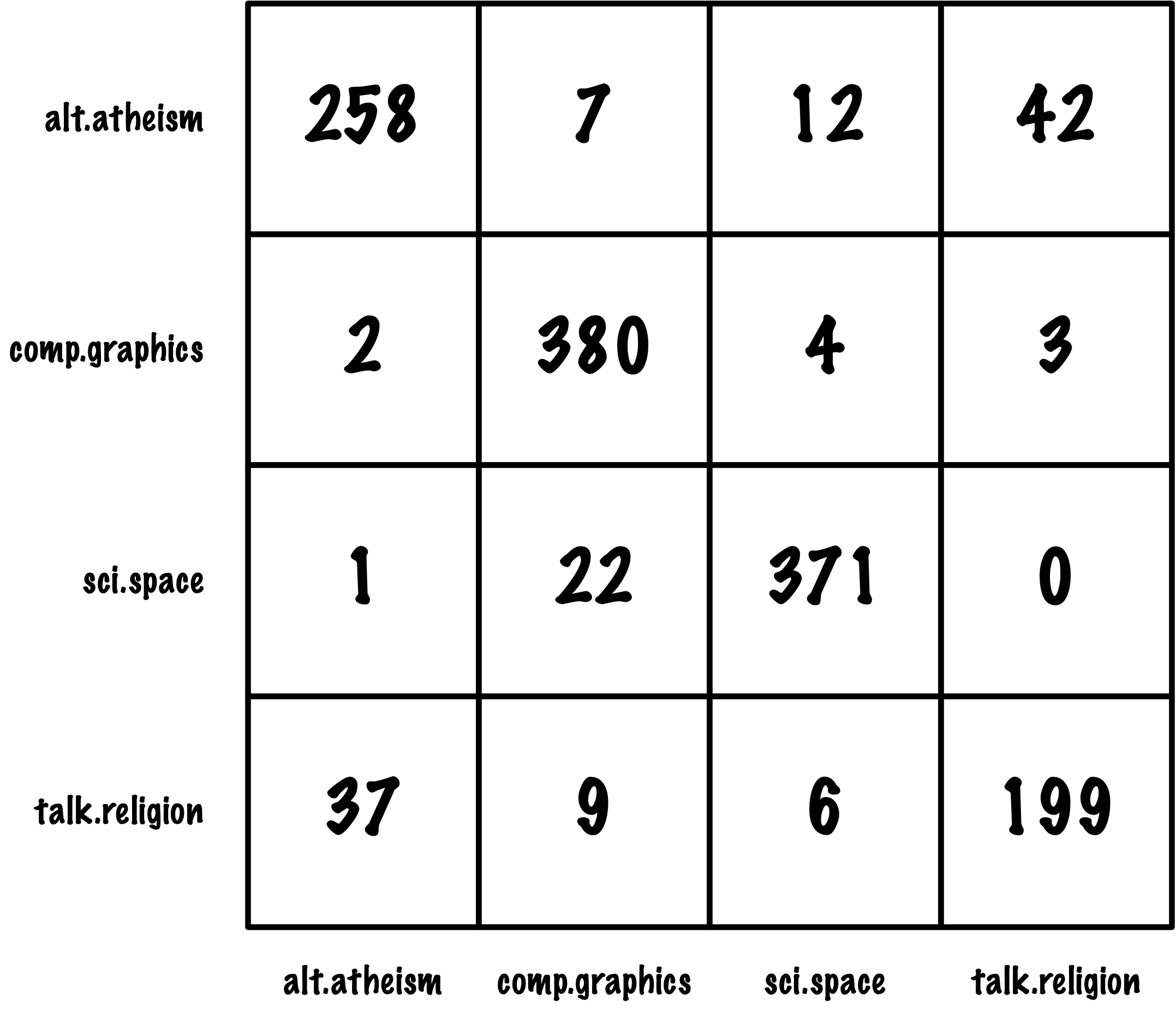
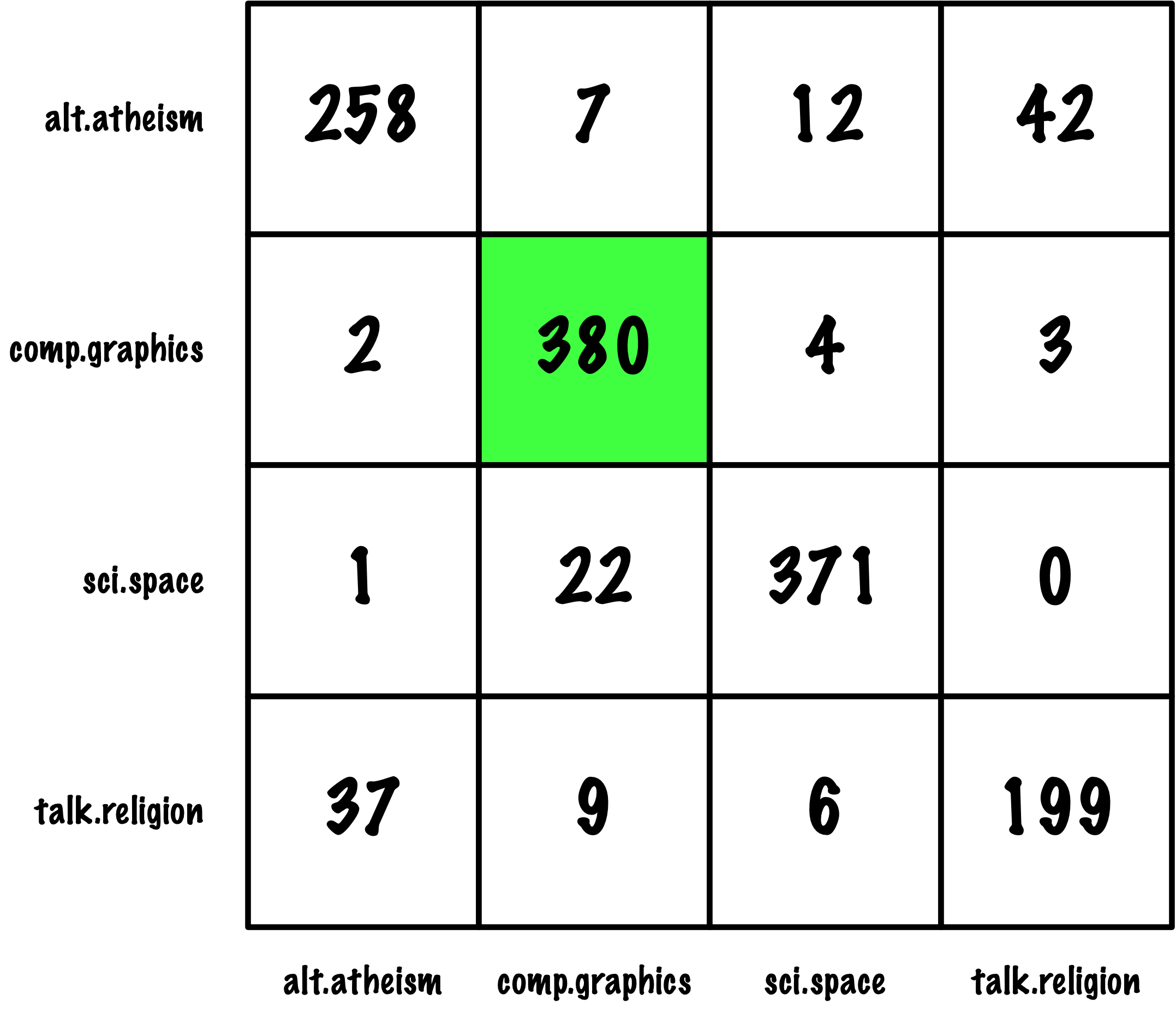
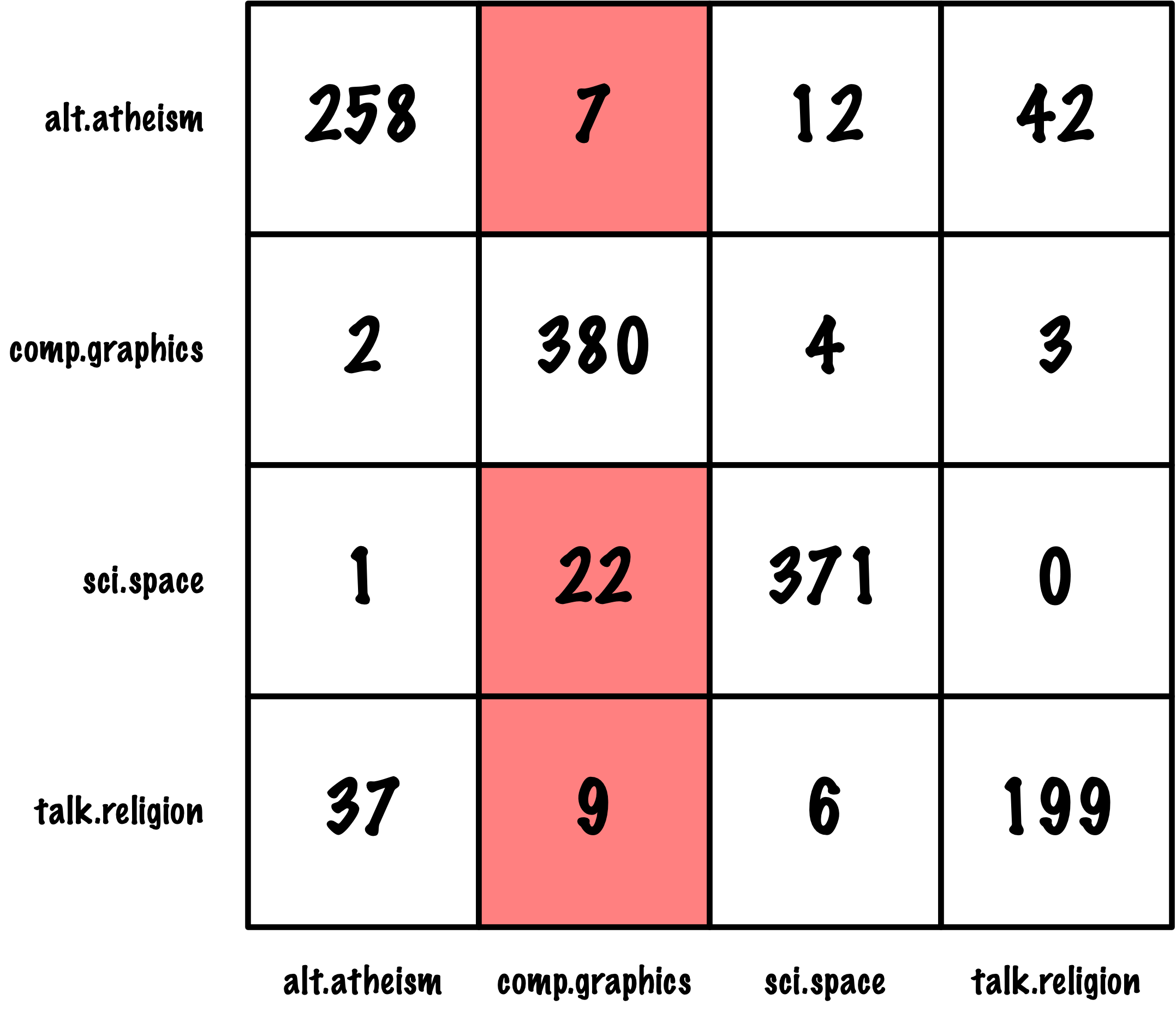
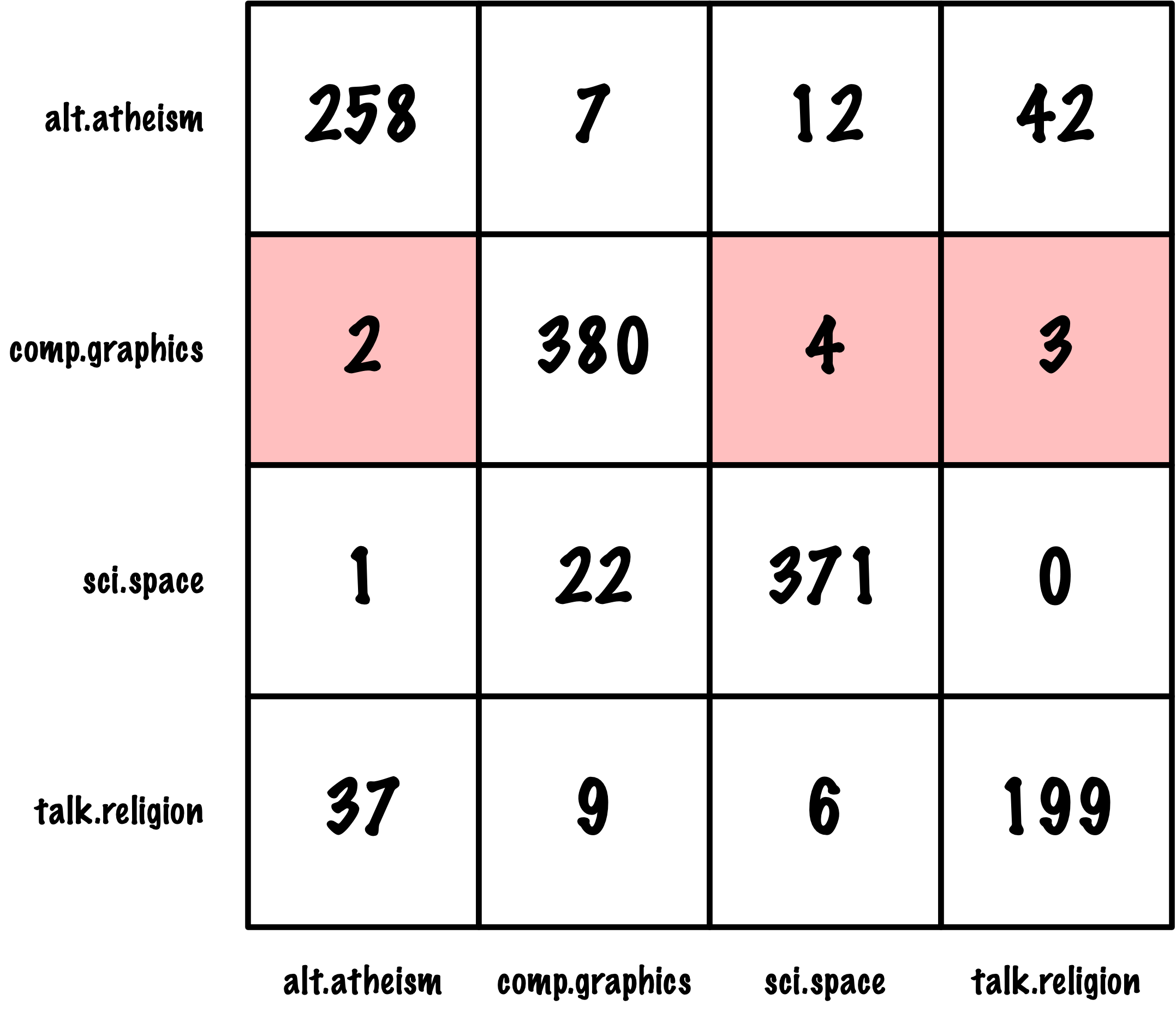
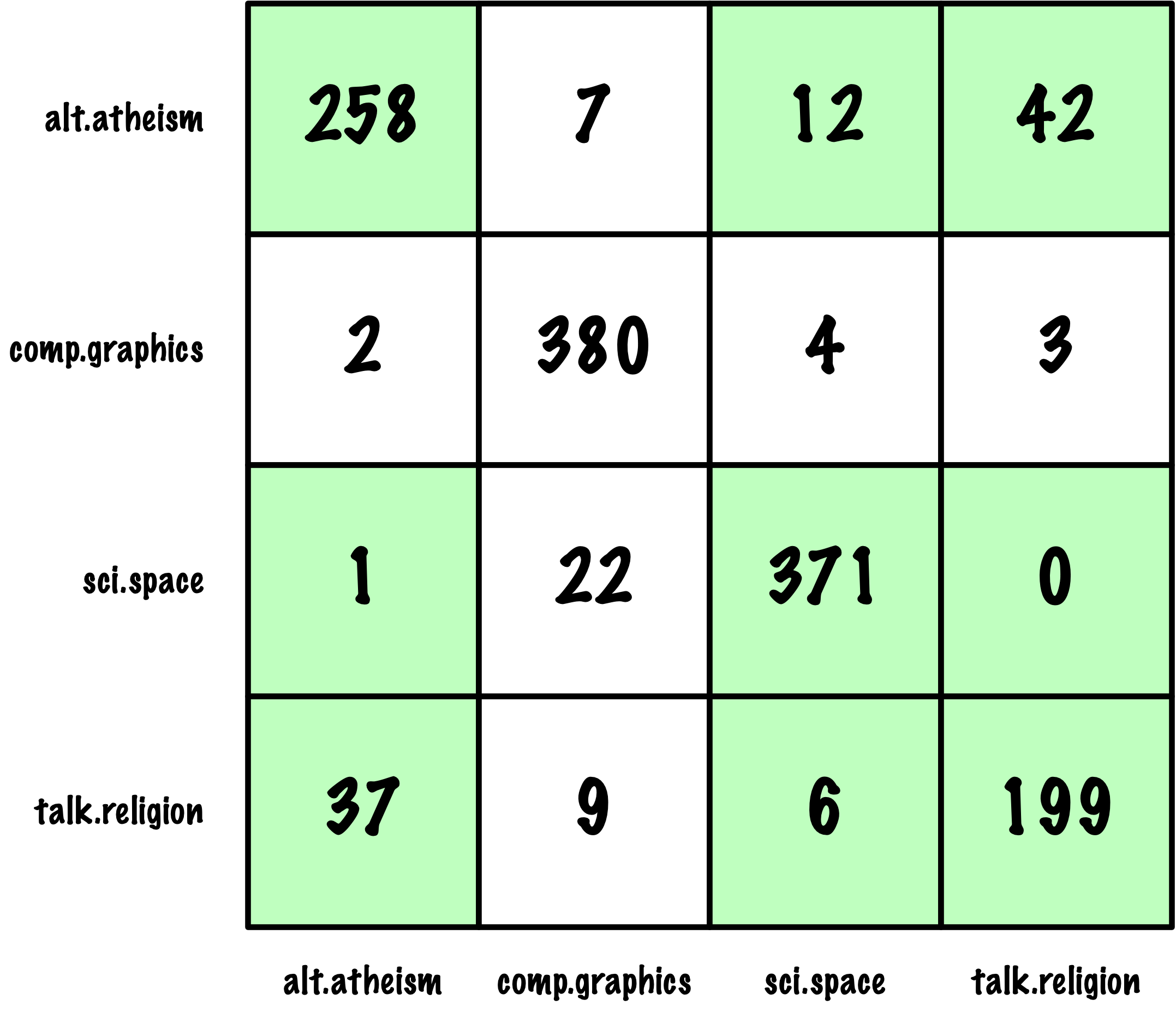
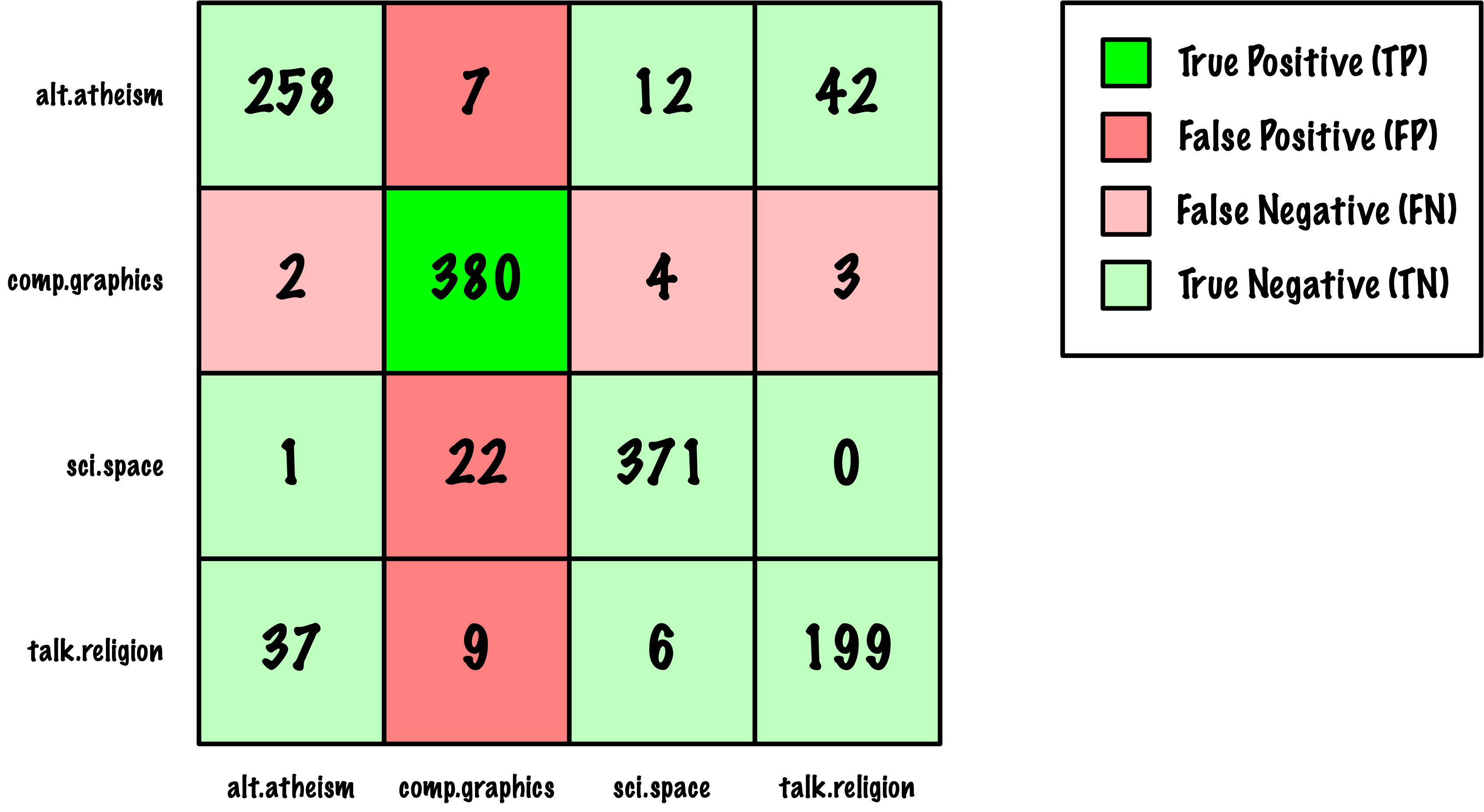
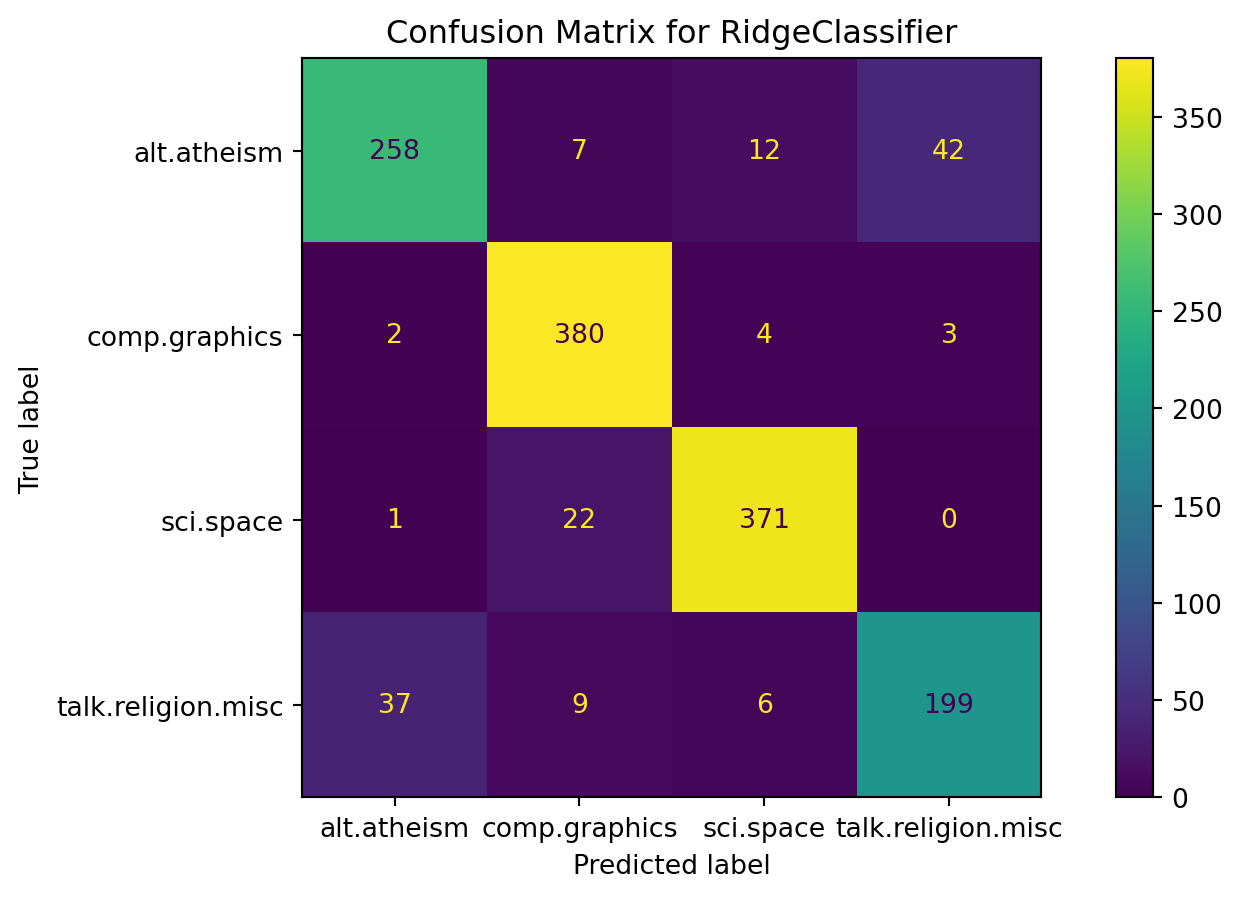
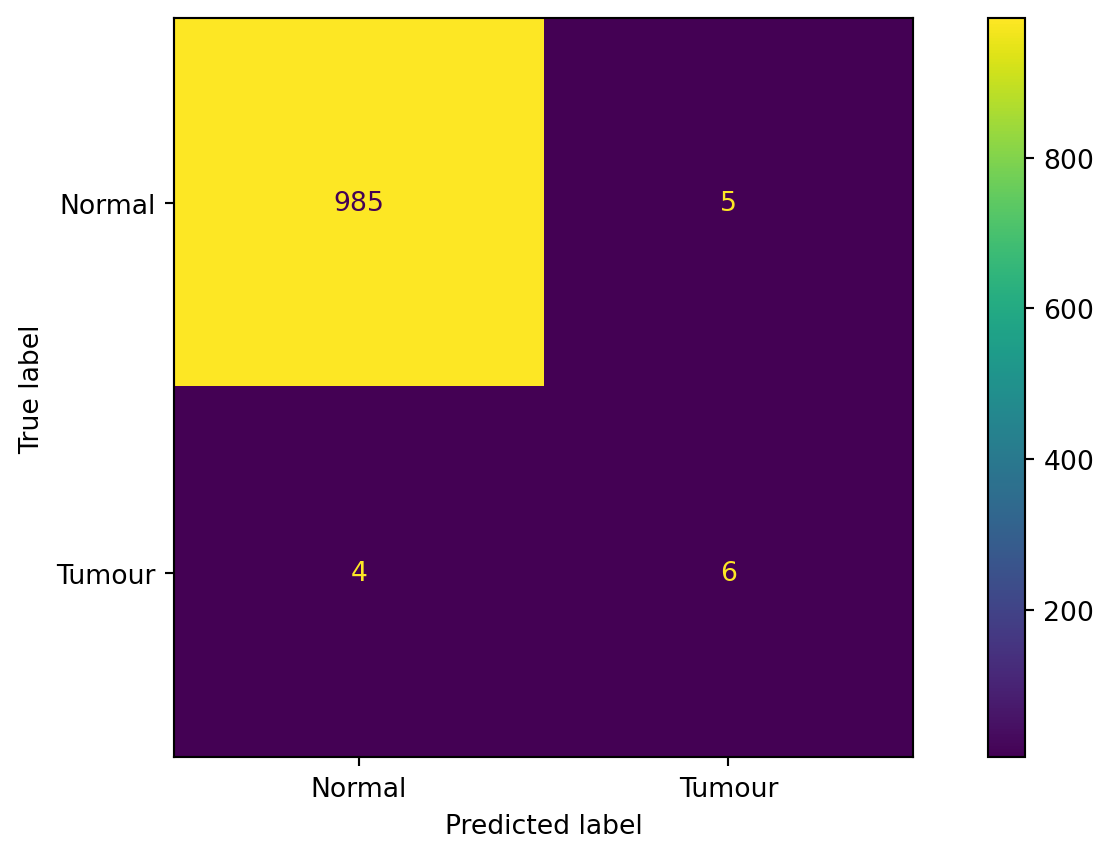
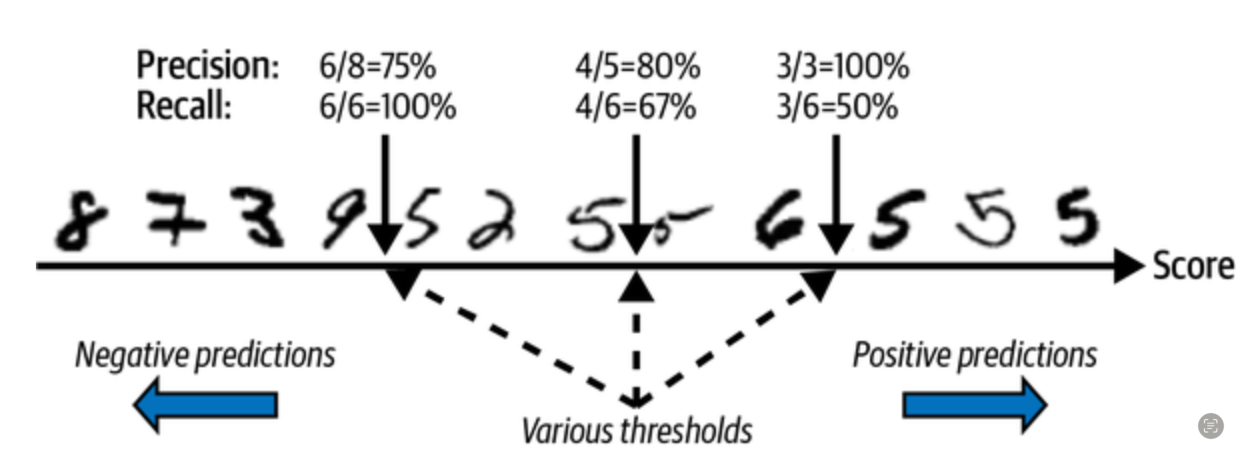
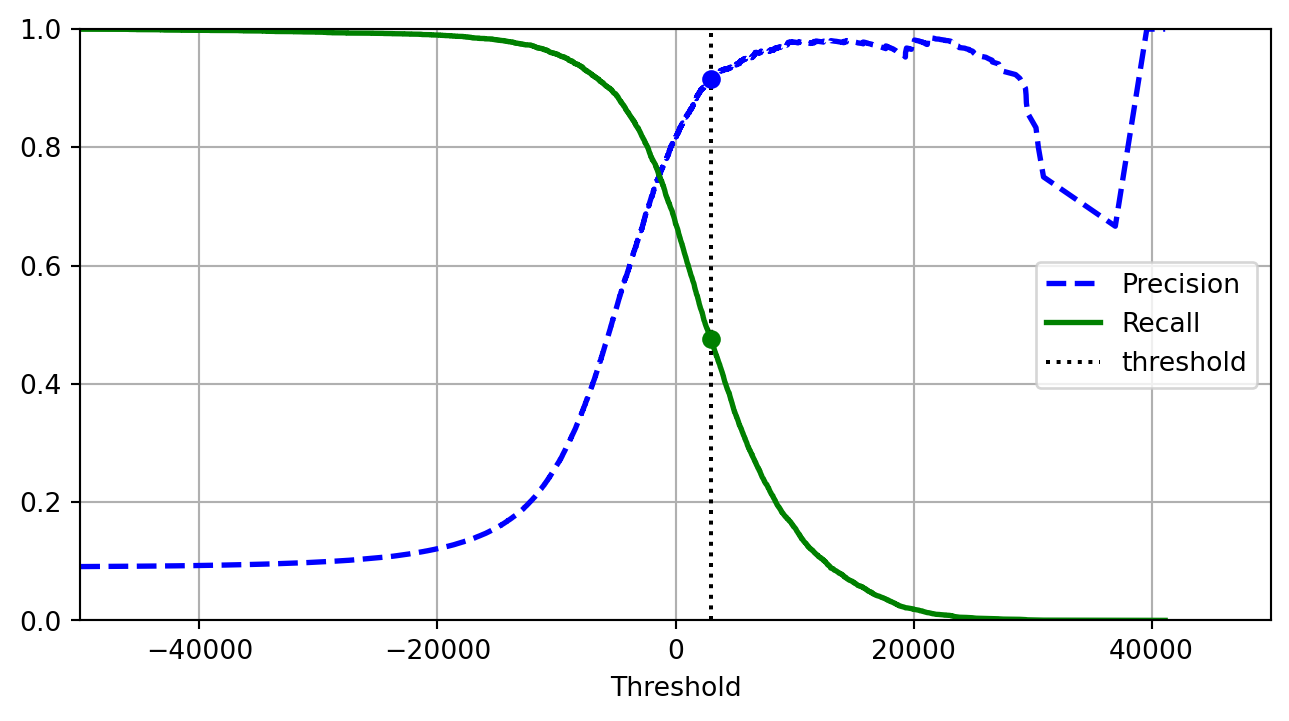
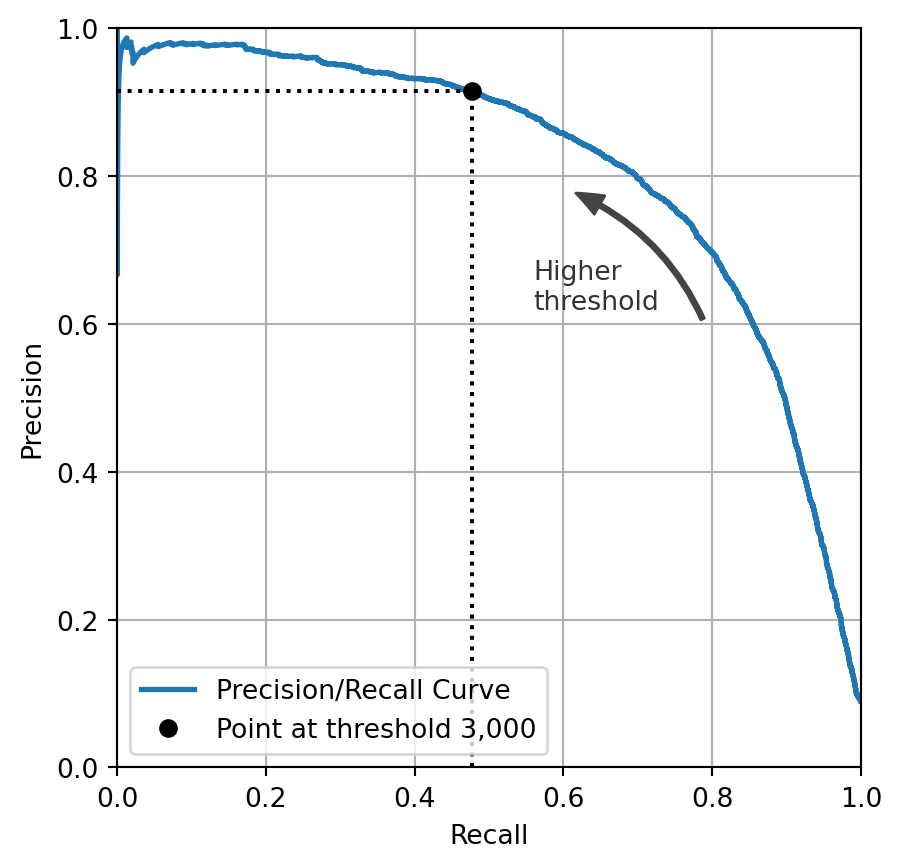
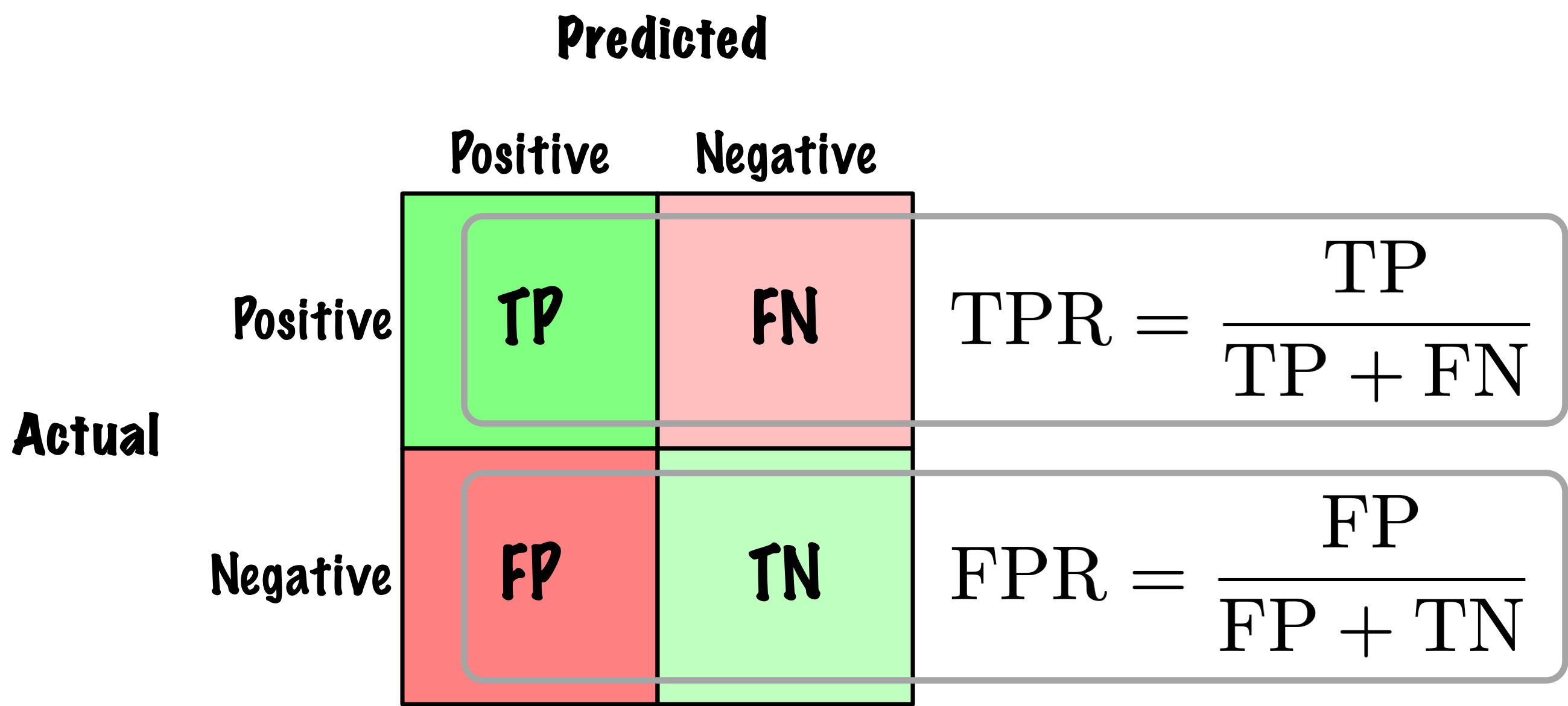
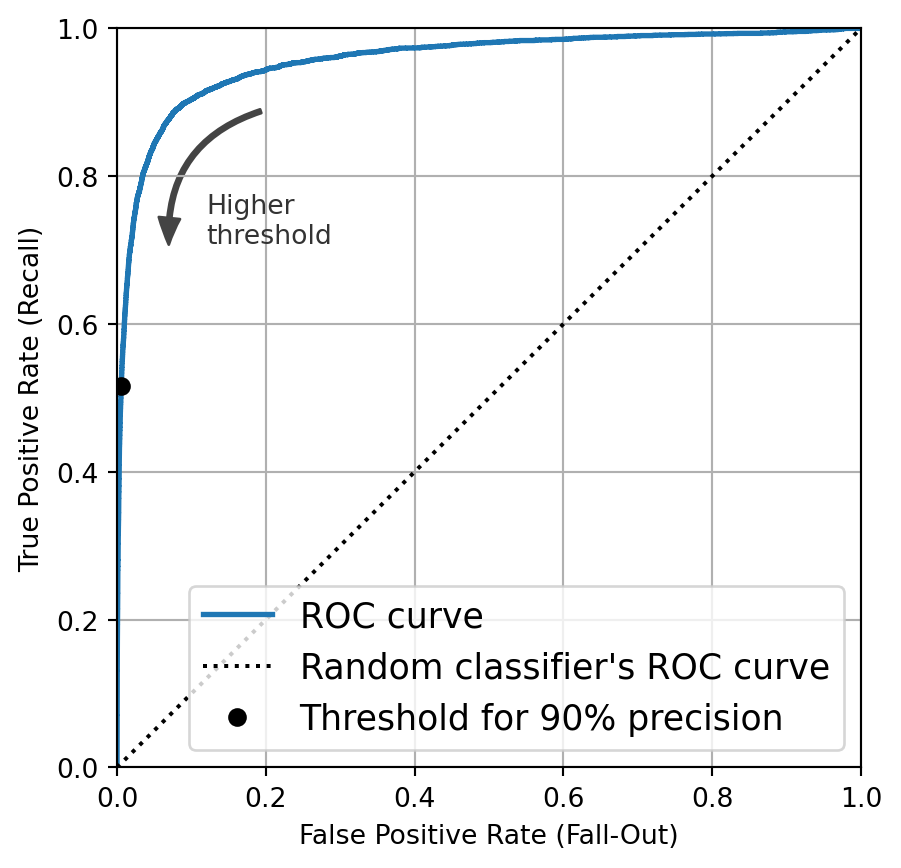
from sklearn.metrics import roc_auc_score
y_pred_prob_lr = lr.predict_proba(X_test)[:, 1]
y_pred_prob_knn = knn.predict_proba(X_test)[:, 1]
y_pred_prob_dt = dt.predict_proba(X_test)[:, 1]
y_pred_prob_rf = rf.predict_proba(X_test)[:, 1]
# Compute ROC curves
fpr_lr, tpr_lr, _ = roc_curve(y_test, y_pred_prob_lr)
fpr_knn, tpr_knn, _ = roc_curve(y_test, y_pred_prob_knn)
fpr_dt, tpr_dt, _ = roc_curve(y_test, y_pred_prob_dt)
fpr_rf, tpr_rf, _ = roc_curve(y_test, y_pred_prob_rf)
# Compute AUC scores
auc_lr = roc_auc_score(y_test, y_pred_prob_lr)
auc_knn = roc_auc_score(y_test, y_pred_prob_knn)
auc_dt = roc_auc_score(y_test, y_pred_prob_dt)
auc_rf = roc_auc_score(y_test, y_pred_prob_rf)
# Plot ROC curves
plt.figure(figsize=(5, 5)) # plt.figure()
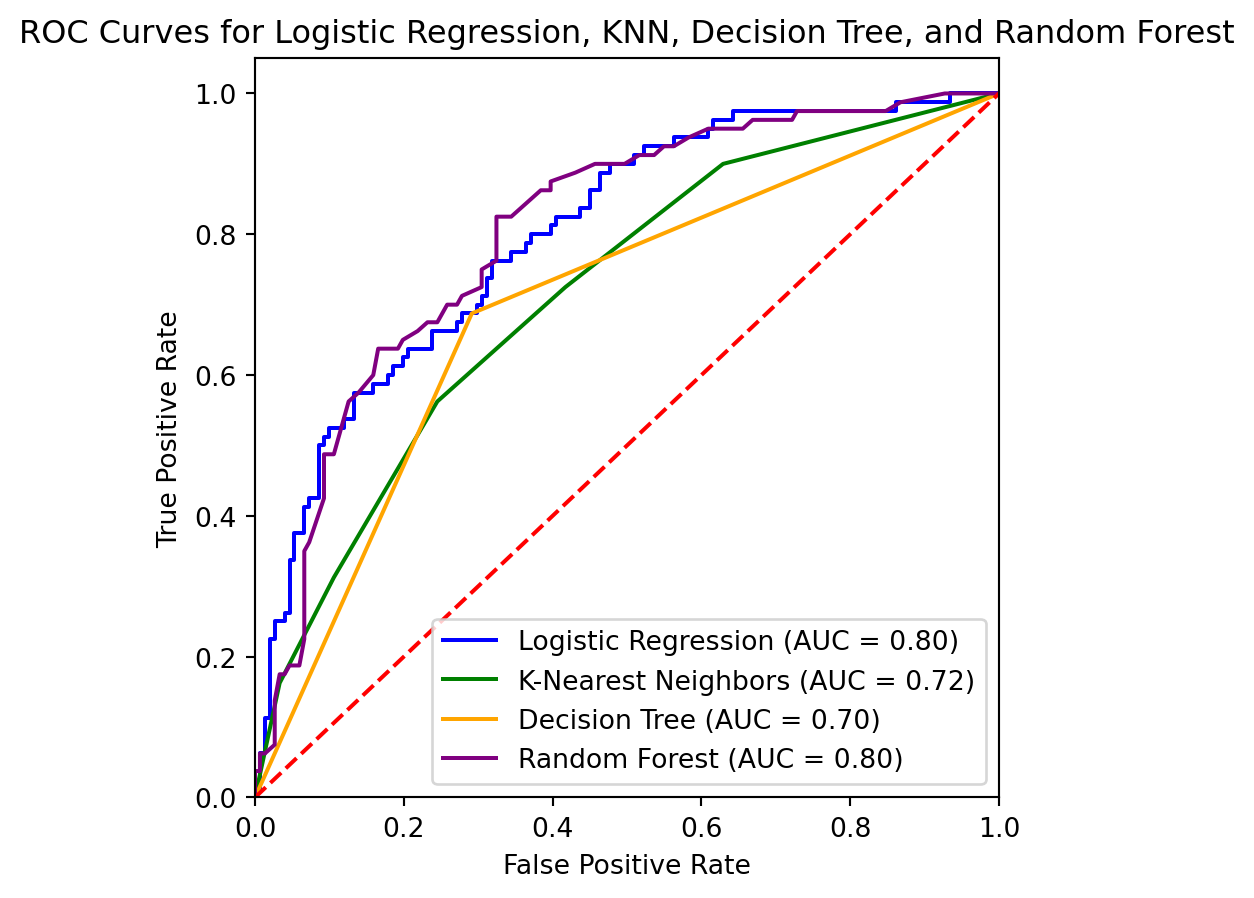
plt.plot(fpr_lr, tpr_lr, color='blue', label=f'Logistic Regression (AUC = {auc_lr:.2f})')
plt.plot(fpr_knn, tpr_knn, color='green', label=f'K-Nearest Neighbors (AUC = {auc_knn:.2f})')
plt.plot(fpr_dt, tpr_dt, color='orange', label=f'Decision Tree (AUC = {auc_dt:.2f})')
plt.plot(fpr_rf, tpr_rf, color='purple', label=f'Random Forest (AUC = {auc_rf:.2f})')
plt.plot([0, 1], [0, 1], color='red', linestyle='--') # Diagonal line for random chance
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ROC Curves for Logistic Regression, KNN, Decision Tree, and Random Forest')
plt.legend(loc="lower right")
plt.show()