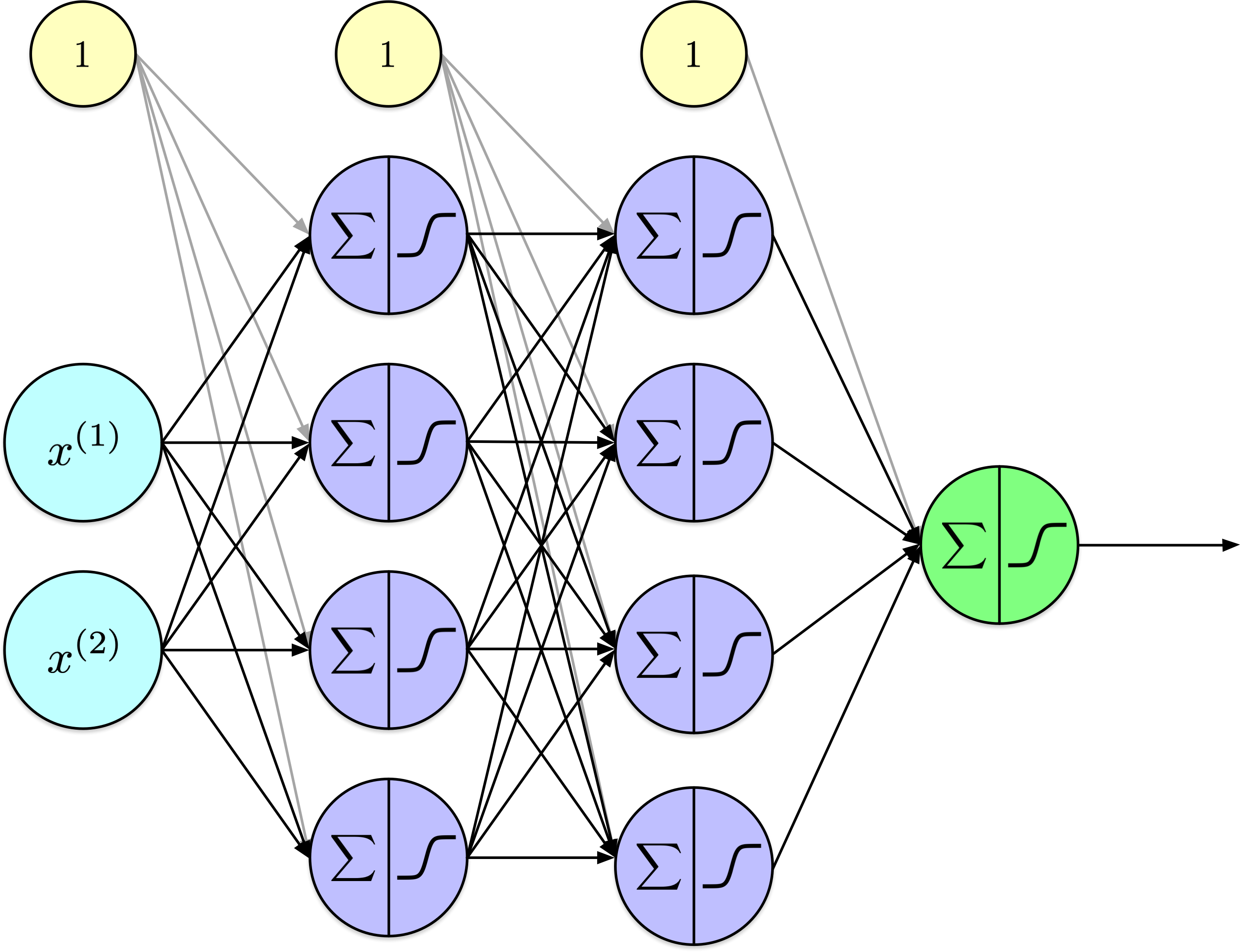
Training Artificial Neural Networks (Part 2)
CSI 4106 - Fall 2025
Version: Oct 26, 2025 11:20
Preamble
Message of the Day
Learning objectives
- Backpropagation Algorithm:
- Discuss the forward and backward passes, highlighting the calculation of gradients using partial derivatives to update weights.
- Vanishing Gradient Problem:
- Outline the issue and present mitigation strategies, such as using activation functions like ReLU or initializing weights with careful consideration.
Back-propagation
Back-propagation
Learning representations by back-propagating errors
David E. Rumelhart, Geoffrey E. Hinton & Ronald J. Williams
We describe a new learning procedure, back-propagation, for networks of neurone-like units. The procedure repeatedly adjusts the weights of the connections in the network so as to minimize a measure of the difference between the actual output vector of the net and the desired output vector. As a result of the weight adjustments, internal ‘hidden’ units which are not part of the input or output come to represent important features of the task domain, and the regularities in the task are captured by the interactions of these units. The ability to create useful new features distinguishes back-propagation from earlier, simpler methods such as the perceptron-convergence procedure.
Before the back-propagation
Limitations, such as the inability to solve the XOR classification task, essentially stalled research on neural networks.
The perceptron was limited to a single layer, and there was no known method for training a multi-layer perceptron.
Single-layer perceptrons are limited to solving classification tasks that are linearly separable.
Back-propagation: contributions
The model employs mean squared error as its loss function.
Gradient descent is used to minimize loss.
A sigmoid activation function is used instead of a step function, as its derivative provides valuable information for gradient descent.
Shows how updating internal weights using a two-pass algorithm consisting of a forward pass and a backward pass.
Enables training multi-layer perceptrons.
Conceptual Idea
Conceptual Idea (continued)
Conceptual Idea (continued)
Backpropagation
Backpropagation is an algorithm for methodically computing the partial derivatives of a neural network’s loss function with respect to each weight and bias parameter.
Backpropagation applies the chain rule of calculus recursively to compute \(\frac{\partial J}{\partial w_{i,j}^{(\ell)}}\) for all network parameters \(w_{i,j}^{(\ell)}\) efficiently, using intermediate quantities from the forward pass, where \(w_{i,j}^{(\ell)}\) denotes the parameter \(w_{i,j}\) of the layer \(\ell\).
Chain rule
Given,
\[ h(x) = f(g(x)) \]
using the Lagrange notation, we have
\[ h^\prime(x) = f^\prime(g(x)) g^\prime(x) \]
or equivalently using Leibniz notation
\[ \frac{dz}{dx} = \frac{dz}{dy} \cdot \frac{dy}{dx} \]
Applying the chain rule recursively
Computational graph
Scalar input; one hidden node
Derivatives
\[ \frac{\partial J}{\partial \hat{y}} \]
\[ \frac{\partial \hat{y}}{\partial z_2} \]
\[ \frac{\partial z_2}{\partial w_2}, \quad \frac{\partial z_2}{\partial b_2}, \quad \frac{\partial z_2}{\partial a_1}, \]
\[ \frac{\partial a_1}{\partial z_1}, \]
\[ \frac{\partial z_1}{\partial w_1}, \quad \frac{\partial z_1}{\partial b_1}, \quad \frac{\partial z_1}{\partial x}, \]
Derivatives
Derivatives
Derivatives
Derivatives
Derivatives
Combined derivatives
For \(w_2\):
\[ \frac{\partial J}{\partial w_2}=\frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial w_2}=\left[-\left(\frac{y}{\hat{y}}-\frac{1-y}{1-\hat{y}}\right)\right] \cdot(\hat{y}(1-\hat{y})) \cdot a_1 \]
Simplifies to:
\[ \frac{\partial J}{\partial w_2}=(\hat{y}-y) a_1 \]
Combined derivatives
Combined derivatives
For \(w_1\):
\[ \frac{\partial J}{\partial w_1}=\frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial a_1} \cdot \frac{\partial a_1}{\partial z_1} \cdot \frac{\partial z_1}{\partial w_1} \]
Plug in:
\[ =\left[-\left(\frac{y}{\hat{y}}-\frac{1-y}{1-\hat{y}}\right)\right] \cdot(\hat{y}(1-\hat{y})) \cdot w_2 \cdot\left(a_1\left(1-a_1\right)\right) \cdot x \]
Simplifies to:
\[ \frac{\partial J}{\partial w_1}=(\hat{y}-y) w_2\left(a_1\left(1-a_1\right)\right) x \]
Combined derivatives
For \(b_1\):
\[ \frac{\partial J}{\partial b_1}=\frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial a_1} \cdot \frac{\partial a_1}{\partial z_1} \cdot \frac{\partial z_1}{\partial b_1} \]
Plug in:
\[ =(\hat{y}-y) w_2\left(a_1\left(1-a_1\right)\right) \cdot 1 \]
Simplifies to:
\[ \frac{\partial J}{\partial b_1}=(\hat{y}-y) w_2\left(a_1\left(1-a_1\right)\right) \]
Key derivatives
\[ \begin{align} \frac{\partial J}{\partial w_2} & = (\hat{y}-y) a_1 \\ \frac{\partial J}{\partial b_2} & = \hat{y}-y \\ \frac{\partial J}{\partial w_1} & = (\hat{y}-y) w_2\left(a_1\left(1-a_1\right)\right) x \\ \frac{\partial J}{\partial b_1} & = (\hat{y}-y) w_2\left(a_1\left(1-a_1\right)\right) \\ \end{align} \]
Exploration
Forward
Backward
def backward():
global alpha, w1, b1, w2, b2, a1, z1, z2, y, y_hat
grad_J_w2 = (y_hat - y) * a1
grad_J_b2 = y_hat - y
grad_J_w1 = (y_hat - y) * w2 * (a1 * (1-a1)) * x
grad_J_b1 = (y_hat - y) * w2 * (a1 * (1-a1))
w2 = w2 - alpha * grad_J_w2
b2 = b2 - alpha * grad_J_b2
w1 = w1 - alpha * grad_J_w1
b1 = b1 - alpha * grad_J_b1Training
Key derivatives
\[ \begin{align} \frac{\partial J}{\partial w_2} & = \frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial w_2}\\ \frac{\partial J}{\partial b_2} & = \frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial b_2}\\ \frac{\partial J}{\partial w_1} & = \frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial a_1} \cdot \frac{\partial a_1}{\partial z_1} \cdot \frac{\partial z_1}{\partial w_1}\\ \frac{\partial J}{\partial b_1} & = \frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \cdot \frac{\partial z_2}{\partial a_1} \cdot \frac{\partial a_1}{\partial z_1} \cdot \frac{\partial z_1}{\partial b_1}\\ \end{align} \]
Key derivatives
Let \[ \begin{align} \delta_1 & = \frac{\partial J}{\partial \hat{y}} \cdot \frac{\partial \hat{y}}{\partial z_2} \\ \delta_2 & = \delta_1 \cdot \frac{\partial z_2}{\partial a_1} \cdot \frac{\partial a_1}{\partial z_1} \end{align} \]
Rewrite \[ \begin{align} \frac{\partial J}{\partial w_2} & = \delta_1 \cdot \frac{\partial z_2}{\partial w_2}\\ \frac{\partial J}{\partial b_2} & = \delta_1 \cdot \frac{\partial z_2}{\partial b_2}\\ \frac{\partial J}{\partial w_1} & = \delta_2 \cdot \frac{\partial z_1}{\partial w_1}\\ \frac{\partial J}{\partial b_1} & = \delta_2 \cdot \frac{\partial z_1}{\partial b_1}\\ \end{align} \]
Backpropagation: top level
(Computational Graph Creation)
Initialization
Forward Pass
Compute Loss
Backward Pass (Backpropagation)
Update the parameters and repeat 3 to 6.
Backpropagation: detailed
(Create the computational graph.)
Initialize the weights and biases.
Forward pass: starting from the input, compute the output of each operation in the graph, and store these values.
Compute loss.
Backward pass: starting from the output and moving backward, for each operation.
Compute the derivative of the output with respect to each of the inputs.
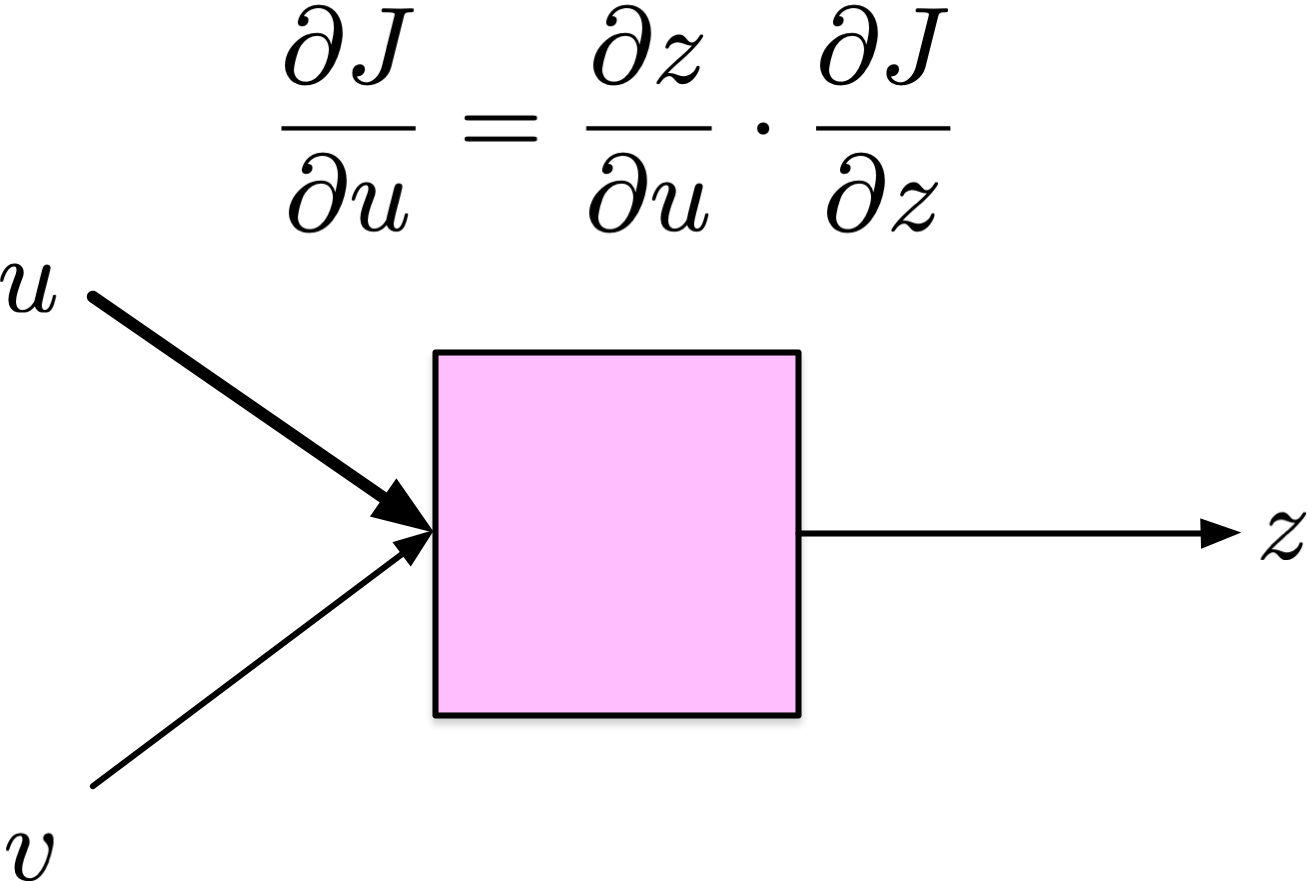
For each input \(u\),
\[ \delta_u = \frac{\partial J}{\partial u} = \frac{\partial z}{\partial u} \cdot \frac{\partial J}{\partial z} \]
- Update the parameters and repeat 3 to 6.
Backpropagation: 2. Initialization
Initialize the weights and biases of the neural network.
Zero Initialization- All weights are initialized to zero.
- Symmetry problems, all neurons produce identical outputs, preventing effective learning.
- Random Initialization
- Weights are initialized randomly, often using a uniform or normal distribution.
- Breaks the symmetry between neurons, allowing them to learn.
- If not scaled properly, leads to slow convergence or vanishing/exploding gradients.
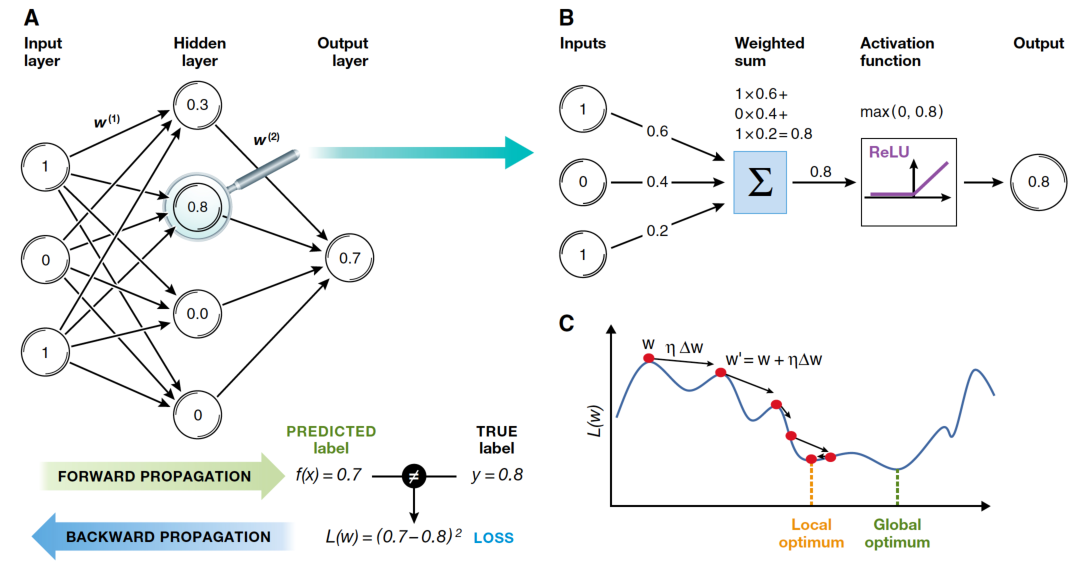
Backpropagation: 3. Forward Pass
For each example in the training set (or in a mini-batch):
Input Layer: Pass input features to first layer.
Hidden Layers: For each hidden layer, compute the activations (output) by applying the weighted sum of inputs plus bias, followed by an activation function (e.g., sigmoid, ReLU).
Output Layer: Same process as hidden layers. Output layer activations represent the predicted values.
Backpropagation: 4. Compute Loss
Calculate the loss (error) using a suitable loss function by comparing the predicted values to the actual target values.
Backpropagation: 5. Backward Pass
Output Layer: Compute the gradient of the loss with respect to the output layer’s weights and biases using the chain rule of calculus.
Hidden Layers: Propagate the error backward through the network, layer by layer. For each layer, compute the gradient of the loss with respect to the weights and biases. Use the derivative of the activation function to help calculate these gradients.
Update Weights and Biases: Adjust the weights and biases using the calculated gradients and a learning rate, which determines the step size for each update.
Backpropagation - Purpose
- Algorithm to train multi-layer perceptrons (MLPs) by minimizing a loss function.
- Enables hidden layers to learn useful internal representations by adjusting weights and biases.
Backpropagation - Core Idea
Iteratively adjust parameters to reduce the difference between predicted and true outputs.
Uses gradient descent on the loss function:
\[ \theta \leftarrow \theta - \alpha \nabla_\theta J(\theta) \]
where \(\alpha\) is the learning rate.
Backpropagation - Summary
Create the computational graph
Initialize weights & biases
Forward pass: compute activations & loss.
Backward pass: compute gradients using chain rule.
Update parameters:
\(W^{(\ell)} \leftarrow W^{(\ell)} - \alpha \frac{\partial J}{\partial W^{(\ell)}}\)
\(b^{(\ell)} \leftarrow b^{(\ell)} - \alpha \frac{\partial J}{\partial b^{(\ell)}}\)
Repeat until convergence.
Implementation
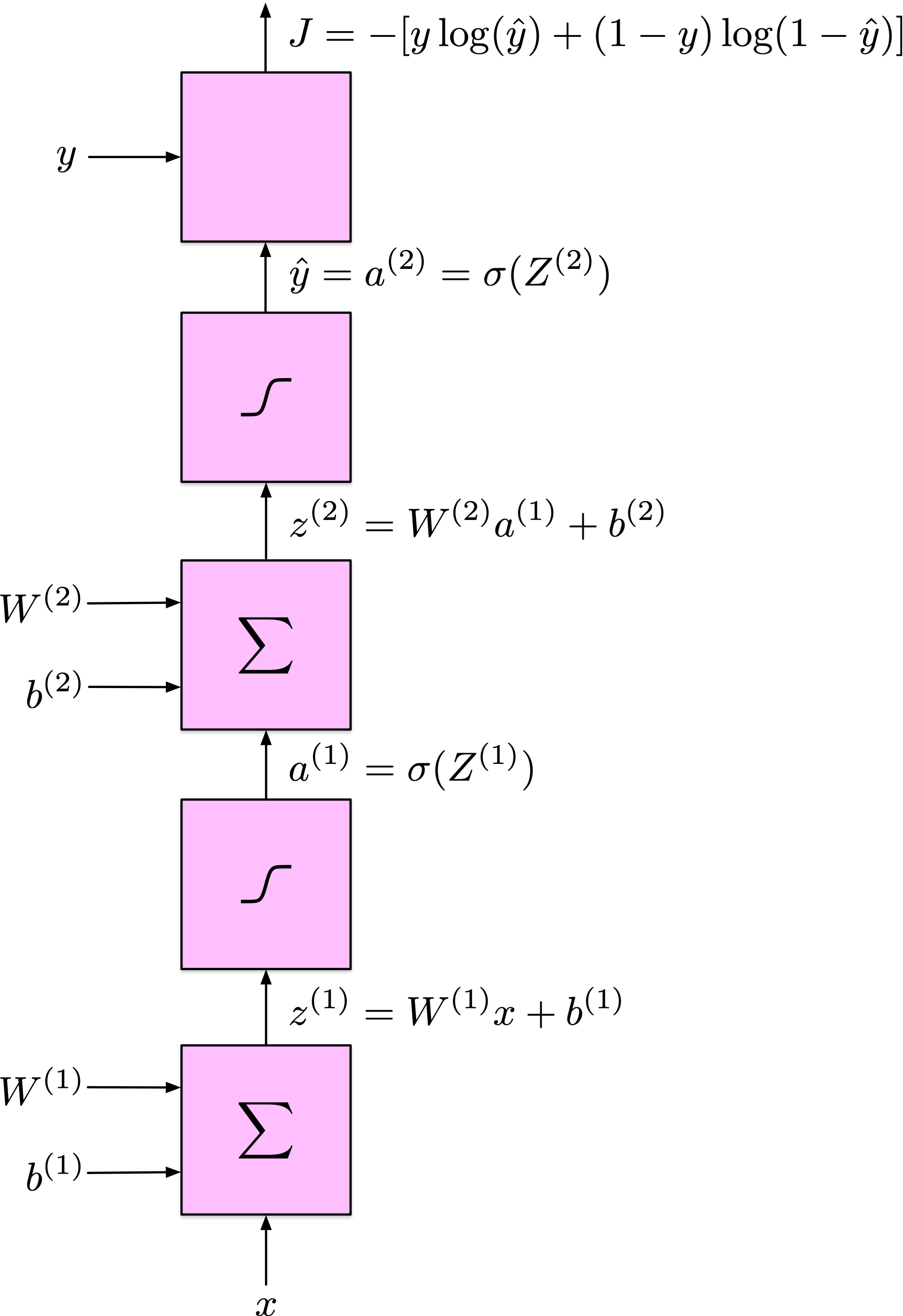
Backpropagation - Forward Pass
Compute activations layer by layer:
\(z^{(\ell)} = W^{(\ell)} a^{(\ell-1)} + b^{(\ell)}\),
\(a^{(\ell)} = \phi(z^{(\ell)})\).
Obtain prediction \(\hat{y}\) and compute loss \(J(\hat{y}, y)\).
Backpropagation - Backward Pass
- Apply chain rule to compute partial derivatives of the loss with respect to each parameter efficiently.
- Propagate gradients from output to input layers:
\[ \delta^{(\ell)} = (W^{(\ell+1)} \delta^{(\ell+1)}) \odot \phi'(z^{(\ell)}) \]
SimpleMLP
The complete implementation is presented below and will be examined in the subsequent slides.
Show code
import numpy as np
# Activations & loss
def sigmoid(z):
return 1.0 / (1.0 + np.exp(-z))
def sigmoid_prime(z):
s = sigmoid(z)
return s * (1.0 - s)
def relu(z):
return np.maximum(0.0, z)
def relu_prime(z):
return (z > 0).astype(z.dtype)
def bce_loss(y_true, y_prob, eps=1e-9):
"""Binary cross-entropy averaged over samples (with clipping for stability)."""
y_prob = np.clip(y_prob, eps, 1 - eps)
return -np.mean(y_true * np.log(y_prob) + (1 - y_true) * np.log(1 - y_prob))
# Initializers
def he_init(rng, fan_in, fan_out):
# He normal: good for ReLU
std = np.sqrt(2.0 / fan_in)
return rng.normal(0.0, std, size=(fan_in, fan_out))
def xavier_init(rng, fan_in, fan_out):
# Glorot/Xavier normal: good for sigmoid/tanh
std = np.sqrt(2.0 / (fan_in + fan_out))
return rng.normal(0.0, std, size=(fan_in, fan_out))
# SimpleMLP (API mirrors NaiveMLP)
class SimpleMLP:
"""
Minimal MLP for binary classification.
- Hidden: ReLU (default) with He init; or 'sigmoid' with Xavier init
- Output: Sigmoid + BCE (δ_L = a_L - y)
- API: forward -> probas (N,), predict_proba, predict, loss, train
"""
def __init__(self, layer_sizes, lr=0.1, seed=None, l2=0.0,
hidden_activation="relu", lr_decay=None):
"""
layer_sizes: e.g., [2, 4, 4, 1]
lr: learning rate
l2: L2 regularization strength (0 disables)
hidden_activation: 'relu' (default) or 'sigmoid'
lr_decay: optional float in (0,1); multiply lr by this every epoch (e.g., 0.9)
"""
self.sizes = list(layer_sizes)
self.lr = float(lr)
self.base_lr = float(lr)
self.lr_decay = lr_decay
self.l2 = float(l2)
self.hidden_activation = hidden_activation
rng = np.random.default_rng(seed)
# Initialize weights/biases per layer
self.W = []
for din, dout in zip(self.sizes[:-1], self.sizes[1:]):
if hidden_activation == "relu":
Wk = he_init(rng, din, dout)
else:
Wk = xavier_init(rng, din, dout)
self.W.append(Wk)
self.b = [np.zeros(dout) for dout in self.sizes[1:]]
# activations (hidden vs output)
def _act(self, z, last=False):
if last:
return sigmoid(z) # output layer
return relu(z) if self.hidden_activation == "relu" else sigmoid(z)
def _act_prime(self, z, last=False):
if last:
return sigmoid_prime(z) # rarely needed with BCE+sigmoid
return relu_prime(z) if self.hidden_activation == "relu" else sigmoid_prime(z)
# forward (public): returns probabilities (N,)
def forward(self, X):
a = X
L = len(self.W)
for ell, (W, b) in enumerate(zip(self.W, self.b), start=1):
a = self._act(a @ W + b, last=(ell == L))
return a.ravel()
# Aliases to match NaiveMLP
def predict_proba(self, X):
return self.forward(X)
def predict(self, X, threshold=0.5):
return (self.predict_proba(X) >= threshold).astype(int)
def loss(self, X, y):
# BCE + optional L2
p = self.predict_proba(X)
base = bce_loss(y, p)
if self.l2 > 0:
reg = 0.5 * self.l2 * sum((W**2).sum() for W in self.W)
# Normalize reg by number of samples to be consistent with mean loss
base += reg / max(1, X.shape[0])
return base
# internal: forward caches for backprop
def _forward_full(self, X):
a = X
activations = [a]
zs = []
L = len(self.W)
for ell, (W, b) in enumerate(zip(self.W, self.b), start=1):
z = a @ W + b
a = self._act(z, last=(ell == L))
zs.append(z)
activations.append(a)
return activations, zs
# training: mini-batch gradient descent with backprop
def train(self, X, y, epochs=30, batch_size=None, verbose=True, shuffle=True):
"""
X: (N, d), y: (N,) in {0,1}
batch_size: None -> full-batch; else int
"""
N = X.shape[0]
idx = np.arange(N)
B = N if batch_size is None else int(batch_size)
for ep in range(1, epochs + 1):
if shuffle:
np.random.shuffle(idx)
if self.lr_decay:
self.lr = self.base_lr * (self.lr_decay ** (ep - 1))
base_loss = self.loss(X, y)
for start in range(0, N, B):
sl = idx[start:start+B]
Xb = X[sl]
yb = y[sl].reshape(-1, 1) # (B,1)
# Forward caches
activations, zs = self._forward_full(Xb)
A_L = activations[-1] # (B,1)
Bsz = Xb.shape[0]
# Backprop
# Output layer: BCE + sigmoid => delta_L = (A_L - y)
delta = (A_L - yb) # (B,1)
grads_W = [None] * len(self.W)
grads_b = [None] * len(self.b)
# Last layer grads
grads_W[-1] = activations[-2].T @ delta / Bsz # (n_{L-1}, 1)
grads_b[-1] = delta.mean(axis=0) # (1,)
# Hidden layers: l = L-1 down to 1
for l in range(2, len(self.sizes)):
z = zs[-l] # (B, n_l)
sp = self._act_prime(z, last=False) # (B, n_l)
delta = (delta @ self.W[-l+1].T) * sp # (B, n_l)
grads_W[-l] = activations[-l-1].T @ delta / Bsz # (n_{l-1}, n_l)
grads_b[-l] = delta.mean(axis=0) # (n_l,)
# L2 regularization (add to grads)
if self.l2 > 0:
for k in range(len(self.W)):
grads_W[k] = grads_W[k] + self.l2 * self.W[k]
# Gradient step
for k in range(len(self.W)):
self.W[k] -= self.lr * grads_W[k]
self.b[k] -= self.lr * grads_b[k]
new_loss = self.loss(X, y)
if verbose:
print(f"Epoch {ep:3d} | loss {base_loss:.5f} → {new_loss:.5f} | Δ={base_loss - new_loss:.5f} | lr={self.lr:.4f}")Activation functions
Loss
Initializers
def he_init(rng, fan_in, fan_out):
# He normal: good for ReLU
std = np.sqrt(2.0 / fan_in)
return rng.normal(0.0, std, size=(fan_in, fan_out))
def xavier_init(rng, fan_in, fan_out):
# Glorot/Xavier normal: good for sigmoid/tanh
std = np.sqrt(2.0 / (fan_in + fan_out))
return rng.normal(0.0, std, size=(fan_in, fan_out))Class definition + constructor
class SimpleMLP:
def __init__(self, layer_sizes, lr=0.1, seed=None, l2=0.0,
hidden_activation="relu", lr_decay=None):
self.sizes = list(layer_sizes)
self.lr = float(lr)
self.base_lr = float(lr)
self.lr_decay = lr_decay
self.l2 = float(l2)
self.hidden_activation = hidden_activation
rng = np.random.default_rng(seed)
# Initialize weights/biases per layer
self.W = []
for din, dout in zip(self.sizes[:-1], self.sizes[1:]):
if hidden_activation == "relu":
Wk = he_init(rng, din, dout)
else:
Wk = xavier_init(rng, din, dout)
self.W.append(Wk)
self.b = [np.zeros(dout) for dout in self.sizes[1:]]Forward (public)
Forward (private)
Training
def train(self, X, y, epochs=30, batch_size=None, verbose=True, shuffle=True):
"""
X: (N, d), y: (N,) in {0,1}
batch_size: None -> full-batch; else int
"""
N = X.shape[0]
idx = np.arange(N)
B = N if batch_size is None else int(batch_size)
for ep in range(1, epochs + 1):
if shuffle:
np.random.shuffle(idx)
if self.lr_decay:
self.lr = self.base_lr * (self.lr_decay ** (ep - 1))
base_loss = self.loss(X, y)
for start in range(0, N, B):
sl = idx[start:start+B]
Xb = X[sl]
yb = y[sl].reshape(-1, 1) # (B,1)
# Forward caches
activations, zs = self._forward_full(Xb)
A_L = activations[-1] # (B,1)
Bsz = Xb.shape[0]
# Backprop
# Output layer: BCE + sigmoid => delta_L = (A_L - y)
delta = (A_L - yb) # (B,1)
grads_W = [None] * len(self.W)
grads_b = [None] * len(self.b)
# Last layer grads
grads_W[-1] = activations[-2].T @ delta / Bsz # (n_{L-1}, 1)
grads_b[-1] = delta.mean(axis=0) # (1,)
# Hidden layers: l = L-1 down to 1
for l in range(2, len(self.sizes)):
z = zs[-l] # (B, n_l)
sp = self._act_prime(z, last=False) # (B, n_l)
delta = (delta @ self.W[-l+1].T) * sp # (B, n_l)
grads_W[-l] = activations[-l-1].T @ delta / Bsz # (n_{l-1}, n_l)
grads_b[-l] = delta.mean(axis=0) # (n_l,)
# L2 regularization (add to grads)
if self.l2 > 0:
for k in range(len(self.W)):
grads_W[k] = grads_W[k] + self.l2 * self.W[k]
# Gradient step
for k in range(len(self.W)):
self.W[k] -= self.lr * grads_W[k]
self.b[k] -= self.lr * grads_b[k]
new_loss = self.loss(X, y)
if verbose:
print(f"Epoch {ep:3d} | loss {base_loss:.5f} → {new_loss:.5f} | Δ={base_loss - new_loss:.5f} | lr={self.lr:.4f}")Testing
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import make_circles
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
X, y = make_circles(n_samples=200, factor=0.5, noise=0.08, random_state=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0, stratify=y)
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
model = SimpleMLP([2, 4, 4, 1], lr=0.3, seed=42, hidden_activation="relu", l2=0.0, lr_decay=0.95)
model.train(X_train, y_train, epochs=150, batch_size=32, verbose=False)
print("Train acc:", accuracy_score(y_train, model.predict(X_train)))
print(" Test acc:", accuracy_score(y_test, model.predict(X_test)))Train acc: 0.95
Test acc: 0.9166666666666666Automatic differentiation
Automatic differentiation (autodiff) systematically applies the chain rule to compute exact derivatives of functions expressed as computer programs. It propagates derivatives through elementary operations, either forward (from inputs to outputs) or backward (from outputs to inputs), enabling efficient and precise gradient computation essential for optimization and learning algorithms.
Training
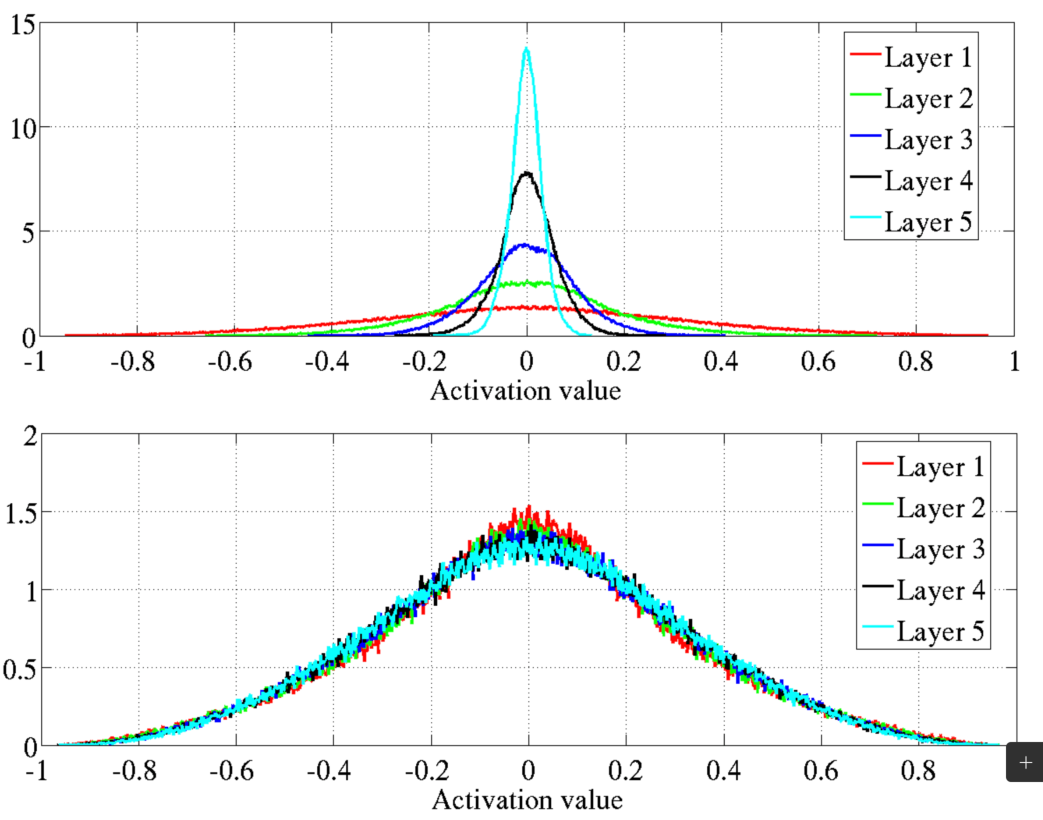
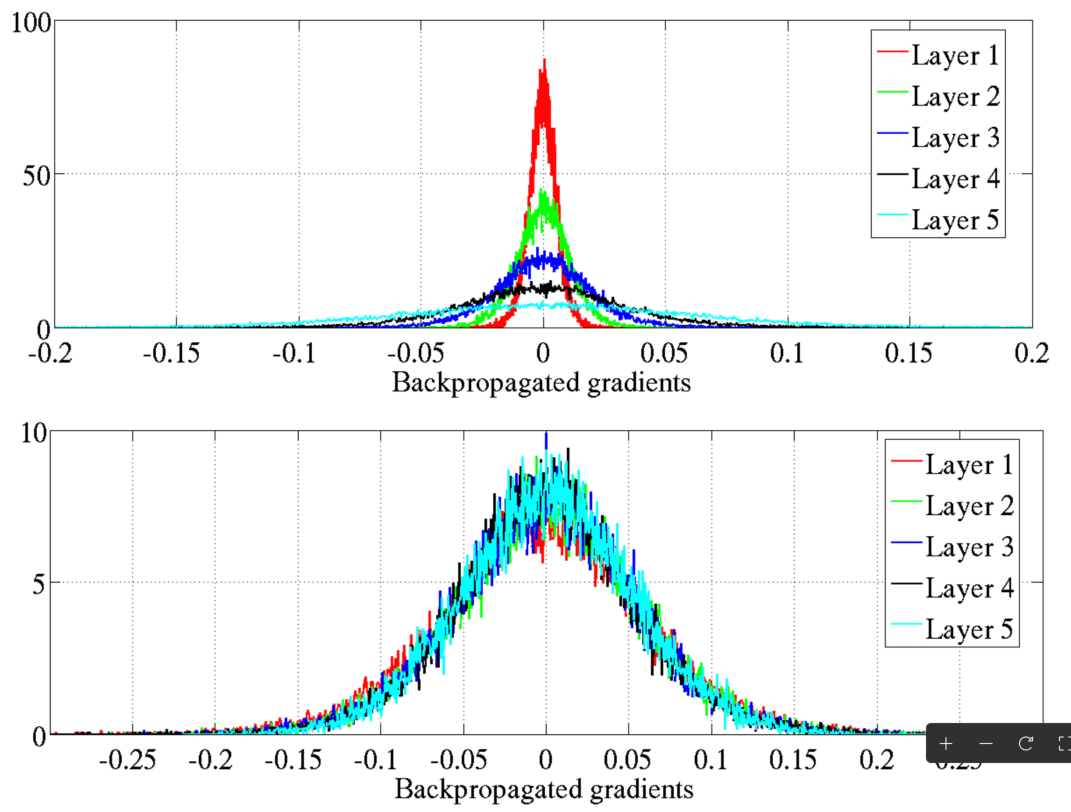
Vanishing gradients
Vanishing gradient problem: Gradients become too small, hindering weight updates.
Stalled neural network research (again) in early 2000s.
Sigmoid and its derivative (range: 0 to 0.25) were key factors.
Common initialization: Weights/biases from \(\mathcal{N}(0, 1)\) contributed to the issue.
Glorot and Bengio (2010) shed light on the problems.
Vanishing gradients: solutions
Alternative activation functions: Rectified Linear Unit (ReLU) and its variants (e.g., Leaky ReLU, Parametric ReLU, and Exponential Linear Unit).
Weight Initialization: Xavier (Glorot) or He initialization.
Glorot and Bengio
He initialization
A similar but slightly different initialization method design to work with ReLU, as well as Leaky ReLU, ELU, GELU, Swish, and Mish.
Note
Randomly initializing the weights1 is sufficient to break symmetry in a neural network, allowing the bias terms to be set to zero without impacting the network’s ability to learn effectively.
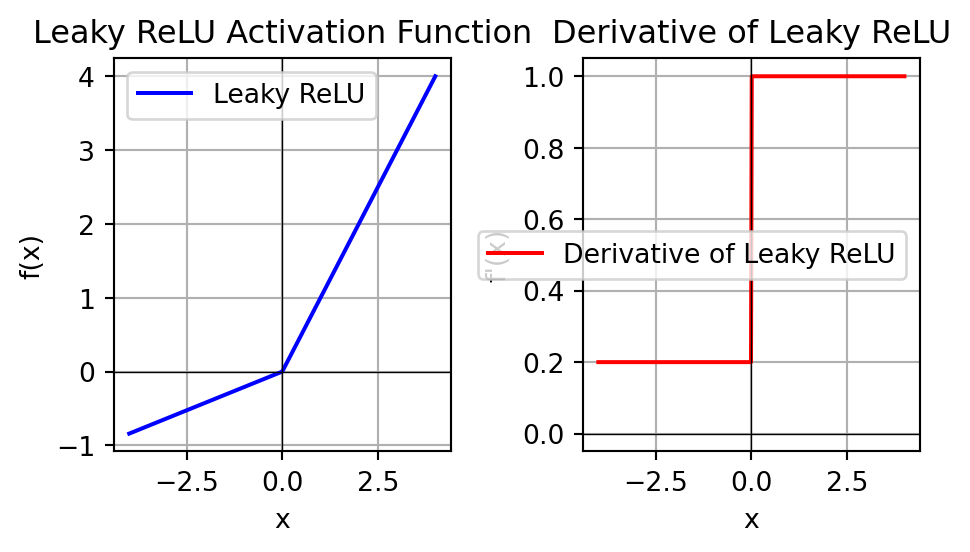
Activation Function: Leaky ReLU
Show code
import numpy as np
import matplotlib.pyplot as plt
# Define the Leaky ReLU function
def leaky_relu(x, alpha=0.21):
return np.where(x > 0, x, alpha * x)
# Define the derivative of the Leaky ReLU function
def leaky_relu_derivative(x, alpha=0.2):
return np.where(x > 0, 1, alpha)
# Generate a range of input values
x_values = np.linspace(-4, 4, 400)
# Compute the Leaky ReLU and its derivative
leaky_relu_values = leaky_relu(x_values)
leaky_relu_derivative_values = leaky_relu_derivative(x_values)
# Create the plot
plt.figure(figsize=(5, 3))
# Plot the Leaky ReLU
plt.subplot(1, 2, 1)
plt.plot(x_values, leaky_relu_values, label='Leaky ReLU', color='blue')
plt.title('Leaky ReLU Activation Function')
plt.xlabel('x')
plt.ylabel('f(x)')
plt.grid(True)
plt.axhline(0, color='black',linewidth=0.5)
plt.axvline(0, color='black',linewidth=0.5)
plt.legend()
# Plot the derivative of the Leaky ReLU
plt.subplot(1, 2, 2)
plt.plot(x_values, leaky_relu_derivative_values, label='Derivative of Leaky ReLU', color='red')
plt.title('Derivative of Leaky ReLU')
plt.xlabel('x')
plt.ylabel("f'(x)")
plt.grid(True)
plt.axhline(0, color='black',linewidth=0.5)
plt.axvline(0, color='black',linewidth=0.5)
plt.legend()
# Show the plots
plt.tight_layout()
plt.show()Prologue
Summary
Summary
| Concept | Role |
|---|---|
| Activation functions | Introduce non-linearity (e.g., Sigmoid, ReLU). |
| Loss function | Measures prediction error (e.g., Binary Cross-Entropy). |
| Learning rate (α) | Controls step size in parameter updates. |
| Gradient descent | Optimization method for weight adjustment. |
| Chain rule | Mechanism for propagating derivatives backward. |
| Automatic differentiation | Software implementation of backprop (e.g., TensorFlow, PyTorch). |
Summary
- Vanishing Gradient Problem:
- Gradients become too small during backpropagation, hindering training.
- Mitigation strategies include using ReLU activation functions and proper weight initialization (Glorot or He initialization).
- Weight Initialization:
- Random initialization breaks symmetry and allows effective learning.
- Glorot initialization suits sigmoid and tanh activations.
- He initialization is optimal for ReLU and its variants.
3Blue1Brown
A series of videos, with animations, providing the intuition behind the backpropagation algorithm.
Neural networks (playlist)
- What is backpropagation really doing? (12m 47s)
- Backpropagation calculus (10m 18s)
StatQuest
Herman Kamper
One of the most thorough series of videos on the backpropagation algorithm.
Introduction to neural networks (playlist)
- Backpropagation (without forks) (31m 1s)
- Backprop for a multilayer feedforward neural network (4m 2s)
- Computational graphs and automatic differentiation for neural networks (6m 56s)
- Common derivatives for neural networks (7m 18s)
- A general notation for derivatives (in neural networks) (7m 56s)
- Forks in neural networks (13m 46s)
- Backpropagation in general (now with forks) (3m 42s)
Free book with implementation
In his book, Neural Networks and Deep Learning, Michael Nielsen provides a comprehensive Python implementation of a neural network.
Next lecture
- We will introduce various architectures of artificial neural networks.
References
Marcel Turcotte
School of Electrical Engineering and Computer Science (EECS)
University of Ottawa