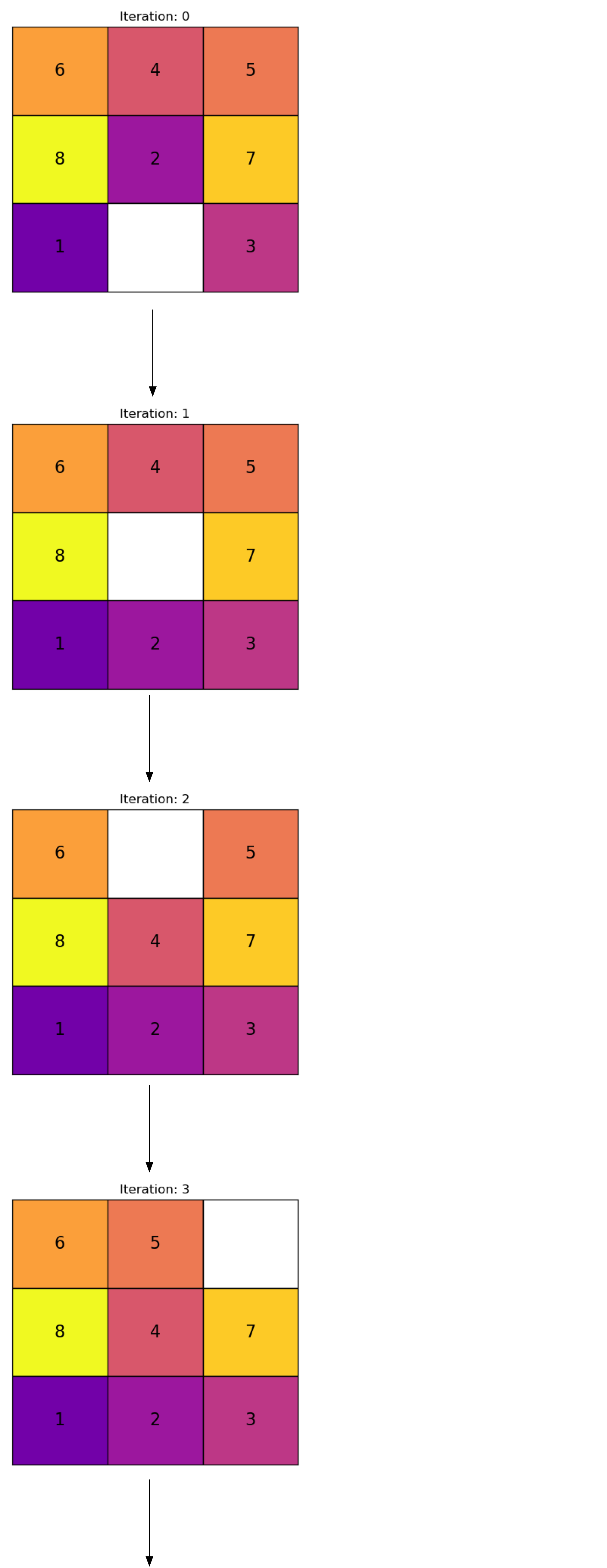
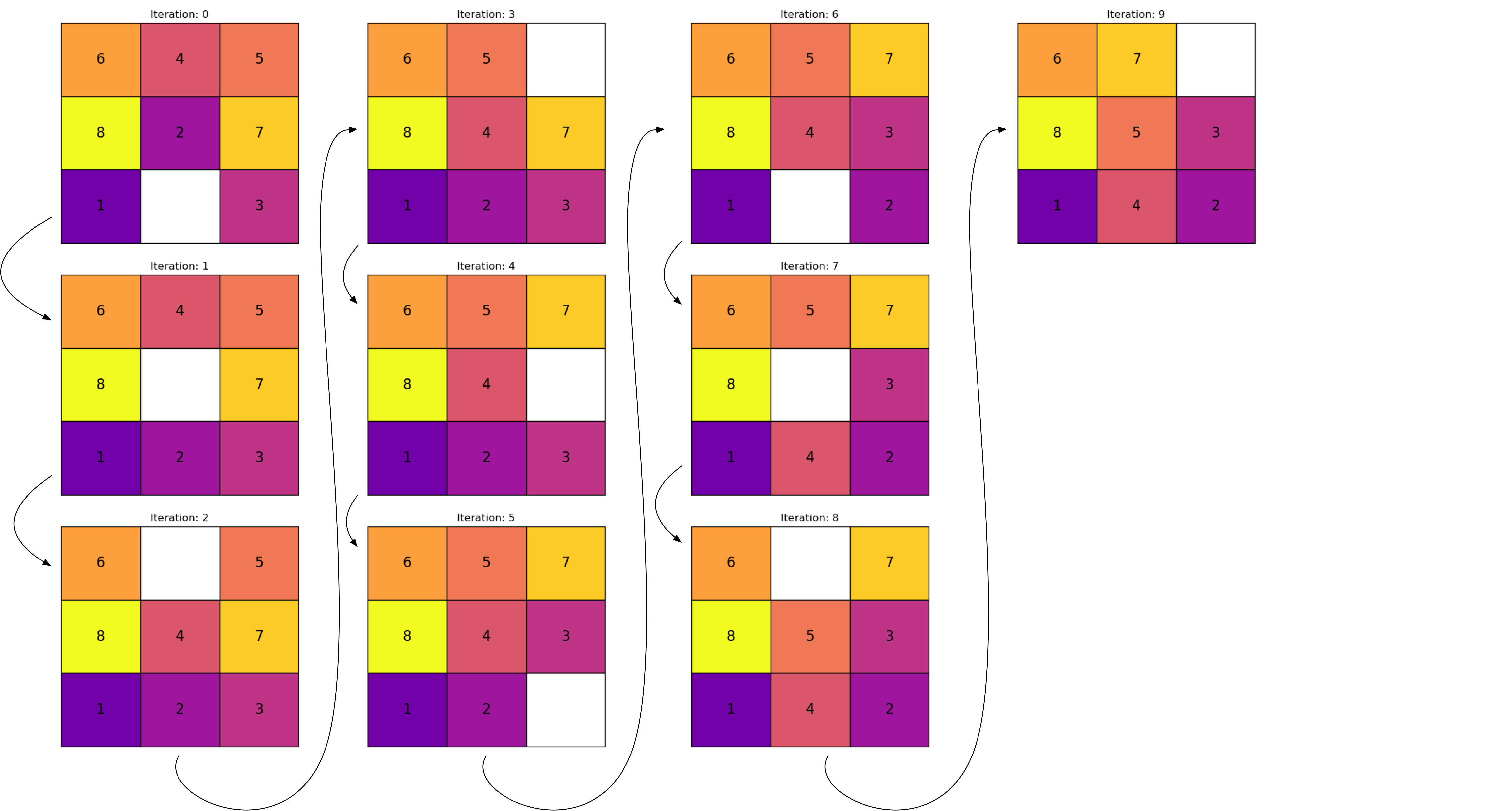
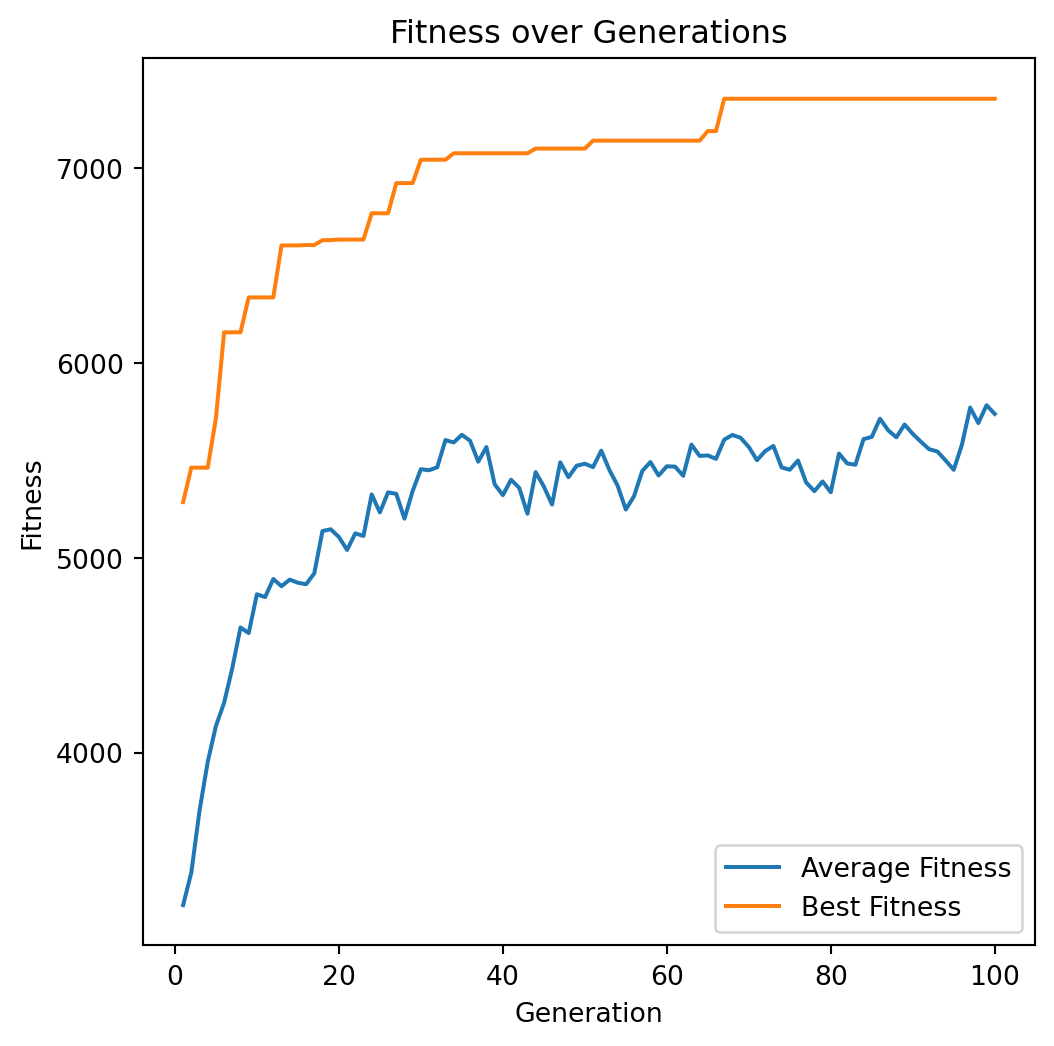
Archer, Aaron F. 1999.
“A Modern Treatment of the 15 Puzzle.” The American Mathematical Monthly 106 (9): 793–99.
https://doi.org/10.1080/00029890.1999.12005124.
Russell, Stuart, and Peter Norvig. 2020.
Artificial Intelligence: A Modern Approach. 4th ed. Pearson.
http://aima.cs.berkeley.edu/.
Schrittwieser, Julian, Ioannis Antonoglou, Thomas Hubert, Karen Simonyan, Laurent Sifre, Simon Schmitt, Arthur Guez, et al. 2020.
“Mastering Atari, Go, chess and shogi by planning with a learned model.” Nature 588 (7839): 604–9.
https://doi.org/10.1038/s41586-020-03051-4.
Silver, David, Aja Huang, Chris J. Maddison, Arthur Guez, Laurent Sifre, George van den Driessche, Julian Schrittwieser, et al. 2016.
“Mastering the game of Go with deep neural networks and tree search.” Nature 529 (7587): 484–89.
https://doi.org/10.1038/nature16961.
Simon, Nisha, and Christian Muise. 2024. “Want To Choose Your Own Adventure? Then First Make a Plan.” Proceedings of the Canadian Conference on Artificial Intelligence.