Essential Cell Biology (Part 2)
CSI 5180 - Machine Learning for Bioinformatics
Version: Jan 20, 2025 09:11
Preamble
Quote of the Day
Previsous Lecture
- Cellular life forms are categorized into two types: prokaryotic and eukaryotic.
- Eukaryotic cells possess organelles, with certain organelles, such as mitochondria, containing their own DNA.
- The three domains of life are Prokarya, Eukarya, and Archaea.
- A phylogeny delineates the evolutionary relationships among organisms and their divergence times.
- The primary macromolecules are DNA, RNA, and proteins.
- Macromolecules are linear, unbranched polymers, wherein each monomer comprises a common backbone and a distinct, specific component, akin to nodes in a linked chain.
Summary
This lecture explores fundamental molecular biology concepts, focusing on the central dogma, the genetic code, and key biological elements: the genome, transcriptome, proteome, and epigenome. The session highlights the significance and application of these concepts in bioinformatics.
General objective
- Describe the central dogma, transcription, translation, and genetic code.
Learning Outcomes
- Central Dogma & Molecular Biology
- Summarize the flow of genetic information (DNA → RNA → Protein).
- Distinguish among replication, transcription, and translation.
- Summarize the flow of genetic information (DNA → RNA → Protein).
- Key Biological Elements
- Differentiate the genome, transcriptome, proteome, and epigenome.
- Recognize how DNA, RNA, and proteins interrelate in gene expression.
- Differentiate the genome, transcriptome, proteome, and epigenome.
- Gene Expression Mechanics
- Identify mRNA, tRNA, rRNA roles and the importance of codons/reading frames.
- Understand promoter regions and the basics of regulatory sequences.
- Identify mRNA, tRNA, rRNA roles and the importance of codons/reading frames.
Decoding a Genomic Revolution
Personalized Medicine
Personalized Medicine involves tailoring medical treatment to the individual characteristics of each patient, including genetic, environmental, and lifestyle factors.
Personalized Medicine
Genetic and Metabolic Profiling: Utilizing an individual’s genetic makeup and metabolic information for therapeutic decisions is a cornerstone of personalized medicine. This approach can lead to better treatment responses and reduced side effects.
Personalized Medicine
Drug Repurposing: Personalized medicine can facilitate drug repurposing by identifying subgroups of patients who may benefit from a drug that has adverse effects in the general population. This can optimize resource use and reduce drug development costs.
Personalized Medicine
“Dimensionally, genetic data is huge,” explains Kim. “Humans have 3.2 billion DNA characters. If you factor in mutations, there are well over 50 million dimensions, and epigenetics is even bigger. Gene expression and transcription — add another 20,000. So put it all together, we’re easily looking at 100-200 million dimensions.
Ultimately, Kim would like to be able to use machine learning to draw all of this genetic, epigenetic and medical record and health data together into a single metric space.
FinnGen
“FinnGen is a research project in genomics and personalized medicine. It is large public-private partnership that has collected and analysed genome and health data from 500,000 Finnish biobank donors to understand the genetic basis of diseases.”
PGP-UK
Note
The Personal Genome Project UK (PGP-UK) is one of few resources that recruits its participants under open consent and makes the resulting multi-omics data freely and openly available.
Proteins
What is a Protein? (from PDB-101)
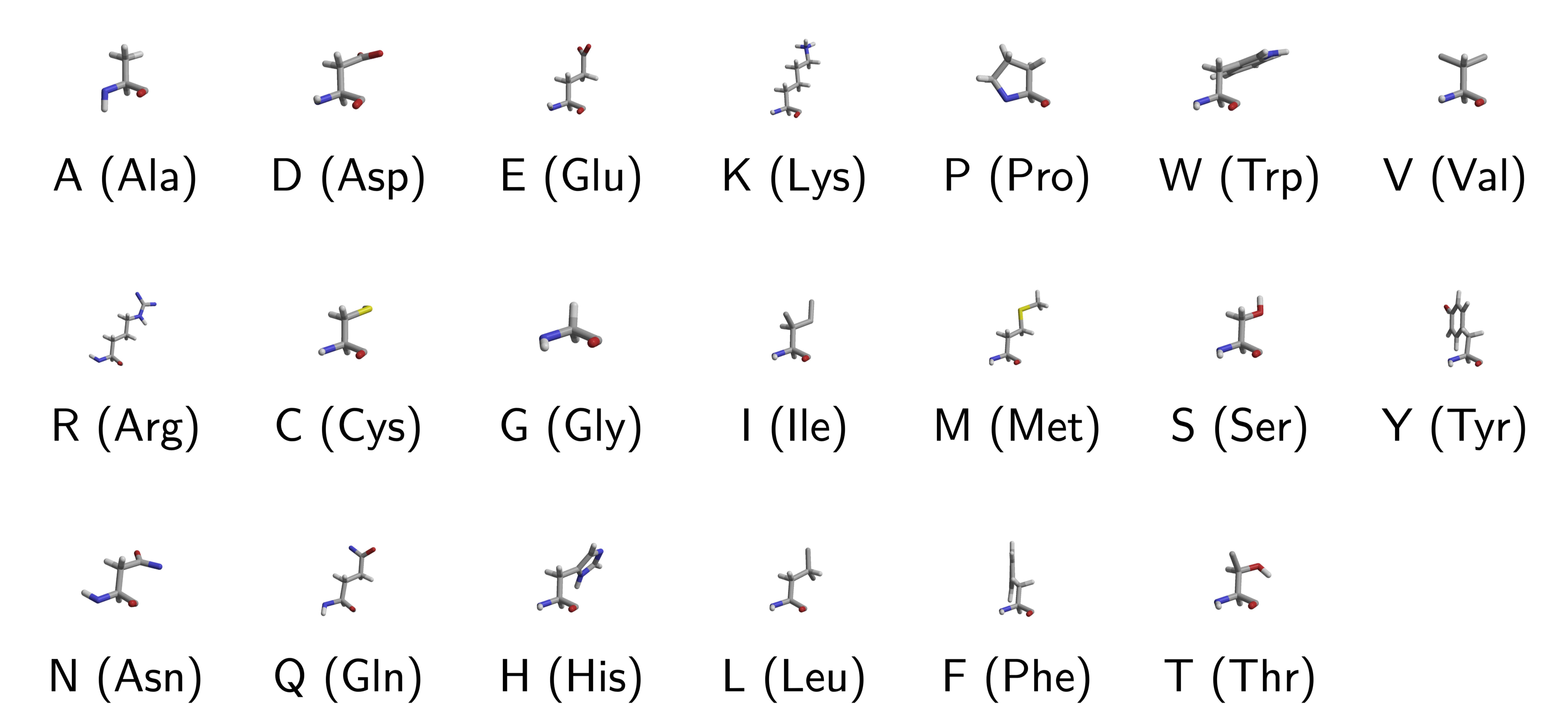
20 (Naturally Occuring) Amino Acids
Genome
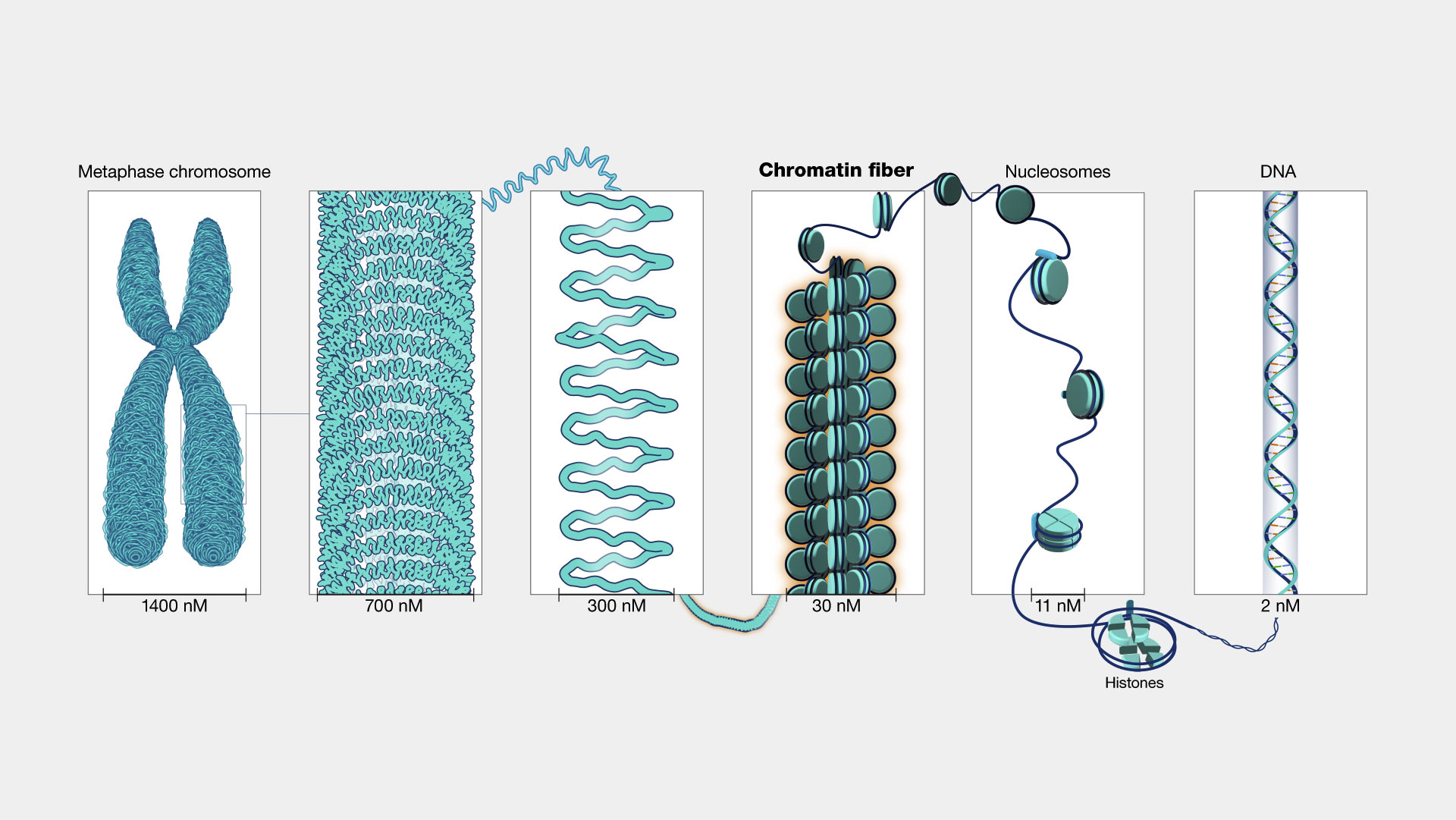
Chromosome Structure
Chromatin
3D Organization of Our Genome (New)
Bioinformaticist’s Perspective
- Predicting histone binding sites based solely on DNA sequence data.
- Utilizing histone locations to infer gene positions and regulatory element sites.
- Exploring the three-dimensional genome organization, a current focal point in research.
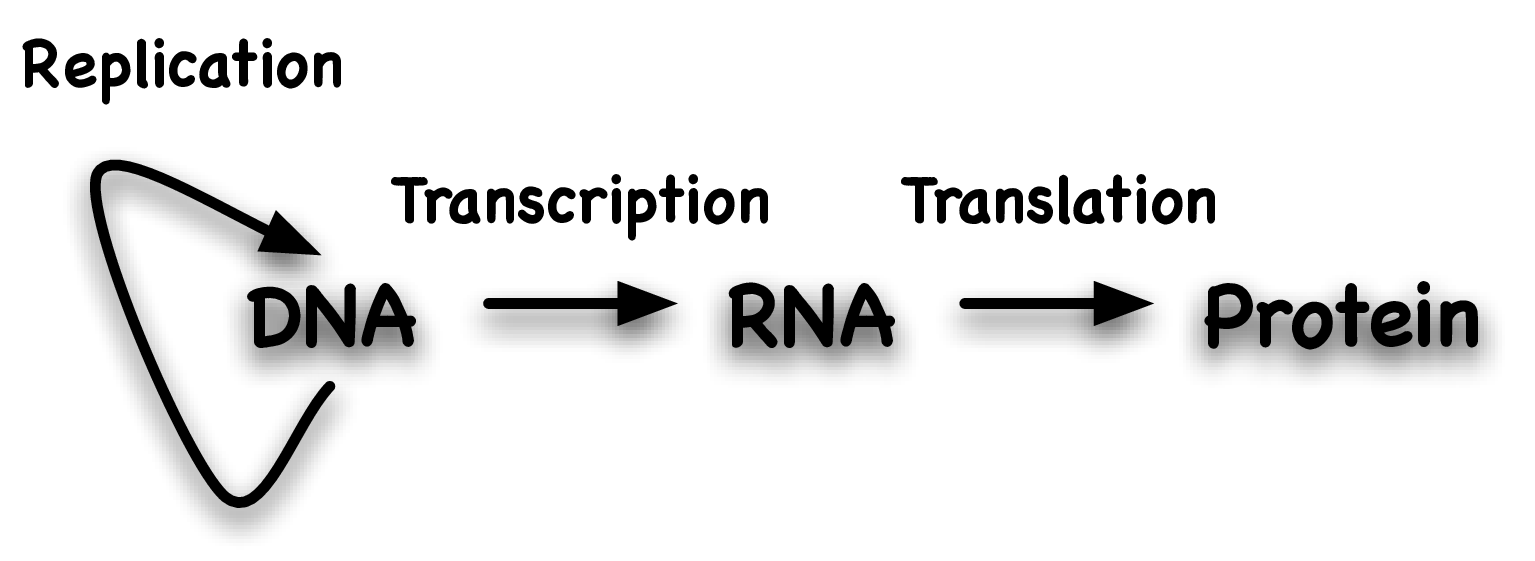
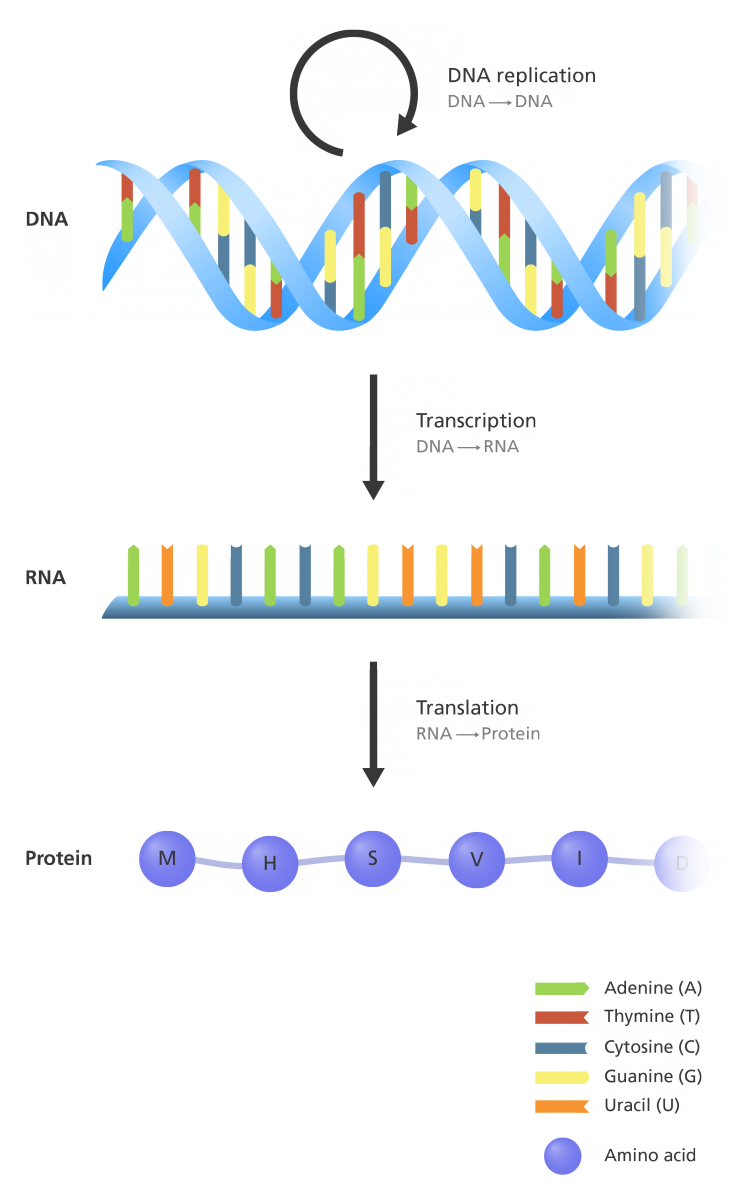
Central Dogma
Central Dogma
DNA, RNA, and proteins are linear sequences of nucleotides or amino acids, representing informational strings.
The Central Dogma of molecular biology elucidates the directional flow of genetic information, delineating the sequential processes by which one macromolecule type dictates the sequence of another.
Fundamentally, this paradigm posits that DNA is transcribed into RNA, which is subsequently translated into protein.
Central Dogma (1958)
Central Dogma (1958)
CRICK (1958)
The central dogma states that once ``information’’ has passed into a protein it cannot get out again. The transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein, may be possible, but transfer from protein to protein, or from protein to nucleic acid, is impossible. Information here means the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.
Central Dogma (today)
Central Dogma (1956)
Central Dogma (DNA)
- DNA: Functions as the repository of genetic information, akin to a comprehensive library of programs.
Central Dogma (RNA)
- RNA: Serves multiple roles including:
- mRNA: Transcribes genetic information for protein synthesis.
- tRNA: Acts as an adaptor in protein synthesis.
- Ribosomal RNA: Integral to ribosomal structure and function.
- Regulatory RNAs: Involved in gene regulation and developmental processes (e.g., microRNAs, riboswitches).
Central Dogma (Proteins)
- Proteins: Perform diverse biological functions, including catalysis, signaling, transport, and structural roles.
Replication
Replication (DNA to DNA)
DNA and Heredity
The structure of DNA elucidates the mechanism by which genetic information is faithfully transmitted across generations or from a parent cell to its daughter cells during the process of replication.
DNA and Heredity (Conceptual)
Before replication
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' BGenerating B’ from A
5' - GATACA -> 3' A
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' B'DNA and Heredity
Before replication
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' BGenerating A’ from B
5' - TGTATC -> 3' B
5' - TGTATC -> 3' B
||||||
3' <- ACATAG -> 5' A'DNA and Heredity
Parent cell AB
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' BDaughter cell AB’
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' B'Daughter cell A’B
5' - TGTATC -> 3' B
||||||
3' <- ACATAG -> 5' A'DNA and Heredity
Parent cell AB
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' BDaughter cell AB’
5' - GATACA -> 3' A
||||||
3' <- CTATGT - 5' B'Daughter cell A’B
5' - GATACA -> 3' A'
||||||
3' <- CTATGT - 5' BRemarks
- Complex organisms develop from a single cell, proliferating into billions of cells. Each cell harbors an identical copy1 of the DNA from its progenitor (parent) cell.
- The redundancy within the DNA double helix allows the second strand’s information to be inferred from the first. This redundancy underpins DNA repair mechanisms, enabling the replacement of deleted bases and the detection of mismatches.
DNA Replication (Basic)
DNA Replication (Advanced)
DNA Replication (Extreme)
Replication: Summary
The process relies critically on the complementarity of base pairs.
Each DNA strand acts as a template for synthesizing its complementary strand.
This results in the formation of two identical double helices, each comprising one parental strand, exemplifying a semi-conservative replication model.
DNA replication is facilitated by the enzyme DNA polymerase.
Observations
Keep these observations in mind as we proceed with the presentation.
- The process of DNA replication is facilitated by various enzymes, including DNA polymerase, Primase, Ligase, and DNA helicase.
- An enzyme is a macromolecule that acts as a catalyst to accelerate specific chemical reactions, with most enzymes being proteins, including those mentioned.
- What is the origin of proteins?
- How are proteins regulated?
Transcription
Transcription (DNA to RNA)
Transcription (Basic)
Genes
Li and Graur (1991)
(\(\ldots\)) a gene is a sequence of genomic DNA (\(\ldots\)) that is essential for a specific function.
There are three (3) kinds of genes:
- Protein-coding genes
- RNA-coding genes
- Regulatory genes
Transcription: DNA to RNA
Necessity of an Intermediate Molecule: In eukaryotic cells, the spatial separation between the nucleus, where DNA resides, and the cytoplasm, where protein synthesis occurs, necessitates the existence of an intermediary molecule to facilitate the transfer of genetic information.
Transcription: DNA to RNA
- Transcription is executed by DNA-dependent RNA polymerase.
- It necessitates specific upstream sequences, termed promoter signals, to initiate transcription of protein-coding genes.
- In eukaryotic organisms, the initial messenger RNA, or pre-mRNA, includes non-coding regions known as introns, which are excised through intron splicing processes.
Transcription (continued)
- In prokaryotes, gene transcription is mediated by a single RNA polymerase.
- In contrast, eukaryotic transcription involves three distinct RNA polymerases: RNA polymerase I transcribes rRNA genes, RNA polymerase II is responsible for protein-coding genes and certain small nuclear RNAs (e.g., U6), and RNA polymerase III transcribes small cytoplasmic RNA genes, including tRNA genes, as well as some small nuclear RNAs.
DNA-RNA Relationship
Transcription (continued)
Transcription involves a straightforward one-to-one correspondence between each nucleotide in the DNA template and the resulting RNA strand. Specifically:
- G pairs with C;
- A pairs with U (instead of T in DNA);
- Utilizes ribonucleotides rather than deoxyribonucleotides.
The resultant molecule is termed a (pre-) messenger RNA or transcript.
Transcription (continued)
- Is the entire genome transcribed?
- No, transcription initiates at specific regions known as promoters.
The consensus sequence for the core promoter in E. coli (Escherichia coli) is as follows:
TTGACA(N){16,18}TATAATWhat is the probability of this motif occurring?
Transcription (continued)
The most straightforward model is the independent and identically distributed (i.i.d.) model.
This model assumes two main principles:
Independence: The probability of the entire motif is the product of the probabilities of each nucleotide at its respective position, indicating no interdependence between positions.
Identical Distribution: The probability distribution for nucleotides remains consistent across all positions in the motif.
Typically, maximum likelihood estimators are employed to determine these probability distributions. This involves gathering extensive sample data and using nucleotide frequency as an estimator for probability.
Promoter Prediction
Consider the canonical promoter motif:
TTGACA(N){16,18}TATAATAssume uniform nucleotide distribution, with probabilities \(p_A = p_C = p_G = p_T = \frac{1}{4}\). Consequently, the likelihood of observing this motif is \(\frac{1}{4^{12}} \approx 6 \times 10^{-8}\).
Estimating Promoter Occurrence in E. coli:
- Given the E. coli genome size of approximately 4.6 Mb, the expected occurrences of this motif are calculated as \(6 \times 10^{-8} \times 4.6 \times 10^6 \approx 0.276\), suggesting fewer than one occurrence.
Promoter Prediction
Comparison with Eukaryotic Genomes:
- Eukaryotic genomes, with sizes often reaching billions of base pairs, exhibit greater promoter complexity, reflecting intricate regulatory needs.
Additional Regulatory Elements:
- Beyond promoters, other regulatory sequences serve as binding sites for transcriptional regulators, facilitating either transcriptional enhancement (positive regulation) or repression (negative regulation).
Bioinformaticist’s Perspective
- The identification of novel regulatory motifs, such as promoters and signaling sequences, remains a vibrant research focus.
About the Animation
Walkthrough of the 1 minute 23 seconds animation.
Transcription factors assemble at a DNA promoter region found at the start of a gene. Promoter regions are characterised by the DNA’s base sequence, which contains the repetition TATATA and for this reason is known as the “TATA box”.
About the Animation
- The TATA box is gripped by the transcription factor TFIID (yellow-brown) that marks the attachment point for RNA polymerase and associated transcription factors. In the middle of TFIID is the TATA Binding Protein subunit, which recognises and fastens onto the TATA box. It’s tight grip makes the DNA kink 90 degrees, which is thought to serve as a physical landmark for the start of a gene.
About the Animation
- A mediator (purple) protein complex arrives carrying the enzyme RNA polymerase II (blue-green). It maneuvers the RNA polymerase into place. Other transcription factors arrive (TFIIA and TFIIB - small blue molecules) and lock into place. Then TFIIH (green) arrives. One of its jobs is to pry apart the two strands of DNA (via helicase action) to allow the RNA polymerase to get access to the DNA bases.
About the Animation
- Finally, the initiation complex requires contact with activator proteins, which bind to specific sequences of DNA known as enhancer regions. These regions can be thousands of base pairs away from the initiation complex. The consequent bending of the activator protein/enhancer region into contact with the initiation-complex resembles a scorpion’s tail in this animation.
About the Animation
- The activator protein triggers the release of the RNA polymerase, which runs along the DNA transcribing the gene into mRNA (yellow ribbon).
About the Animation
- The RNA polymerase unzips a small portion of the DNA helix exposing the bases on each strand. One of the strands acts as a template for the synthesis of an RNA molecule. The base-sequence code is transcribed by matching these DNA bases with RNA subunits, forming a long RNA polymer chain.
Transcription (Detailed)
Transcriptome and Gene Regulation
In prokaryotes, mRNA degradation occurs within minutes post-synthesis, whereas in eukaryotes, it takes several hours.
Regulatory and transport functions are mediated by sequences within the untranslated regions (UTRs) of the mRNA transcript.
Prologue
Central Dogma (Futuristic Animation)
Summary
Central Dogma
DNA → RNA → Protein as the core framework of gene expression.Key Omics
- Genome: Complete genetic blueprint
- Transcriptome: All RNA transcripts
- Proteome: All proteins
- Genome: Complete genetic blueprint
Next Lecture
- Essential Cell Biology (Part 2)
References
Marcel Turcotte
School of Electrical Engineering and Computer Science (EECS)
University of Ottawa