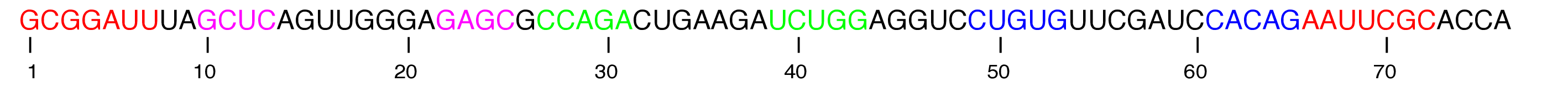Essential Cell Biology (Part 3)
CSI 5180 - Machine Learning for Bioinformatics
Version: Jan 17, 2025 10:51
Preamble
Quote of the Day
The holy grail of pandemic preparedness is being able to predict how a virus will evolve just by looking at its genetic sequence. Those days are still a way off, but a growing number of research groups are using artificial intelligence (AI) to predict the evolution of SARS-CoV-2, influenza and other viruses.
Previsous Lecture
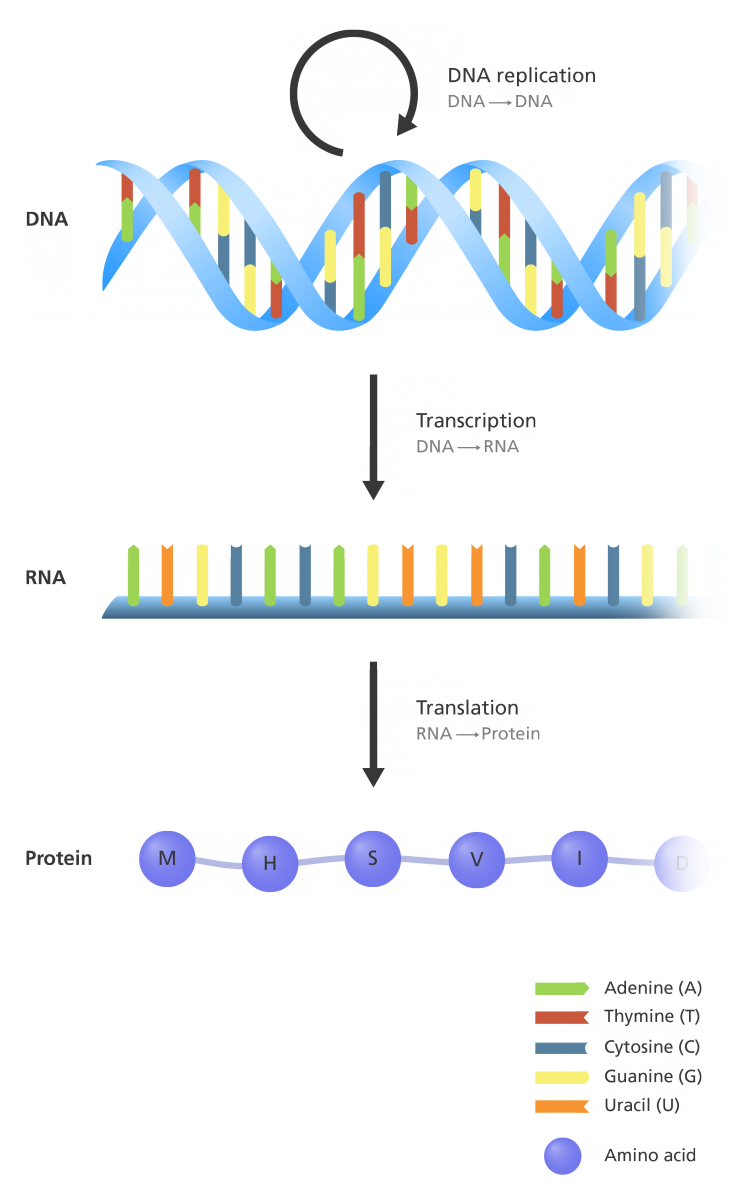
The Central Dogma delineates the flow of genetic information: DNA is transcribed into RNA, which is subsequently translated into proteins.
This process involves three primary stages: replication, transcription, and translation.
Given that DNA resides in the nucleus, while protein synthesis occurs in the cytoplasm, the intermediate molecule messenger RNA (mRNA) is essential for conveying genetic information.
Base pair complementarity is crucial for the fidelity of both replication and transcription processes.
Proteins such as helicase, polymerase, and primase are integral to these processes, facilitating DNA unwinding, strand synthesis, and primer formation, respectively.
Transcription initiation of protein-coding genes is regulated by specific DNA sequences known as promoter signals.
Summary
This lecture examines the latter stages of the central dogma, focusing on translation and its dependence on the genetic code, codons, and tRNA-mediated amino acid delivery. It then shifts to genome organization, highlighting repetitive DNA and its impact on assembly algorithms and disease. The lecture concludes by outlining proteomic concepts and the intricate networks of biological interactions, emphasizing the multifaceted nature of gene expression and regulation.
General objective
- Describe the central dogma, transcription, translation, and genetic code.
Learning Outcomes
- Explain the role of mRNA, tRNA, and ribosomes in protein synthesis.
- Recognize how codons and the degenerate genetic code direct amino acid selection.
- Identify start and stop codons, and interpret open reading frames (ORFs).
- Compare genomic components (exons, introns, repetitive sequences) and their organization.
- Distinguish between the genome, transcriptome, and proteome, and assess their dynamic nature.
- Understand how repetitive elements complicate sequence assembly and associate with disease.
- Recall the importance of protein–protein and protein–DNA interactions in biological networks.
Translation
Translation (RNA to Protein)
Translation (RNA to Protein)
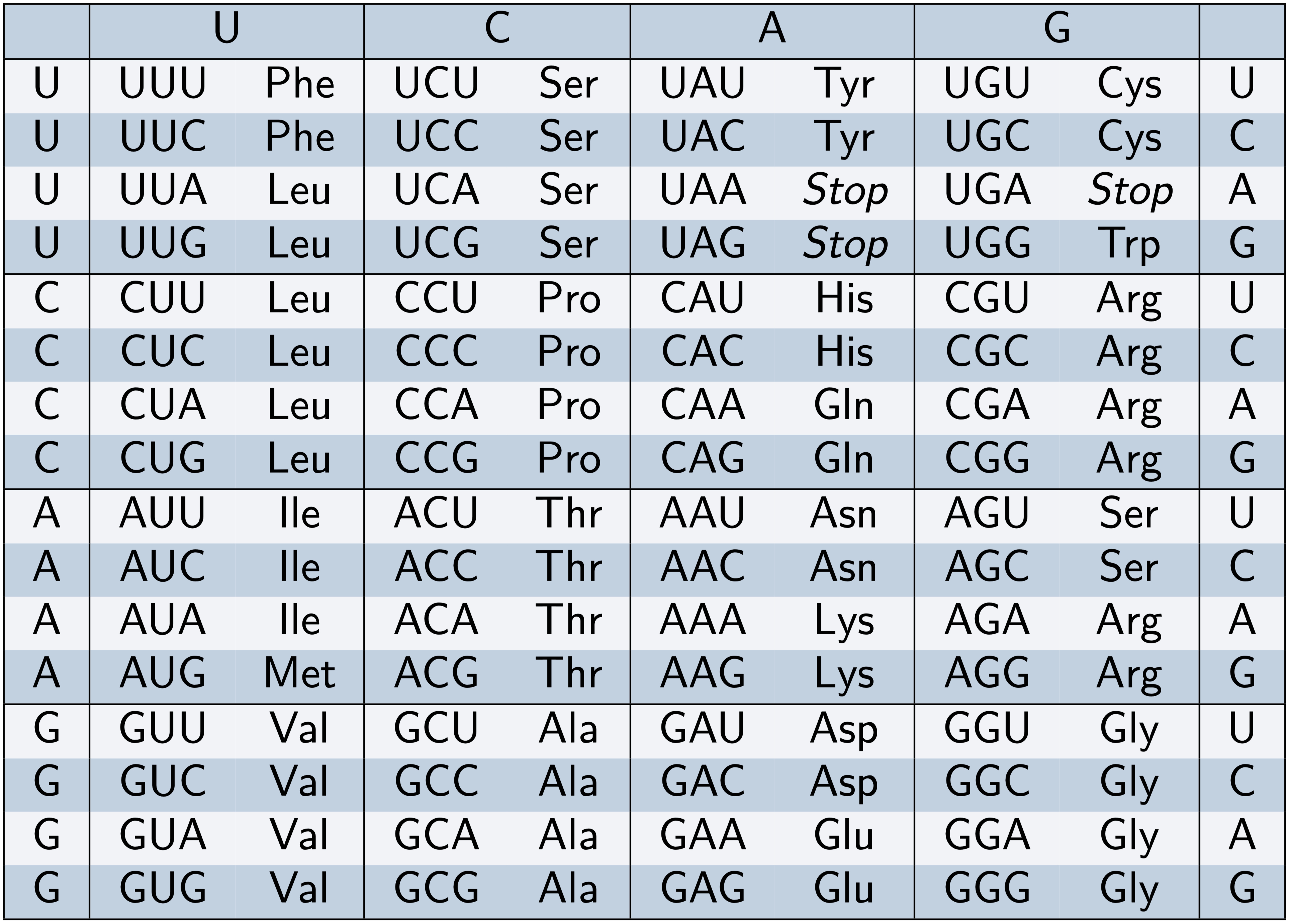
- The genetic code cannot follow a one-to-one mapping:
\[ 4^1 < 20 \]
\[ 4^2 < 20 \]
\[ 4^3 > 20 \]
Translation (RNA to Protein)
- Each set of three consecutive nucleotides forms a codon, which specifies a unique amino acid.
\[ 4^3 = 64 \]
Translation (RNA to Protein)
Codons are arranged in contiguous, non-overlapping triplets.
Given the 64 possible codons, the genetic code is degenerate, meaning that multiple codons can correspond to the same amino acid.
Universal Genetic Code
DNA-RNA-Protein Relationships
DNA: TAC CGC GCC TAT TAC TGC CAG GAA GGA ACT
RNA: AUG GCG CCG AUA AUG ACG GUC CUU CCU UGA
Protein: M A P I M T V L P *
DNA: TAC CGC GCC TAT TAC TGC CAG GAA GGA ACT
RNA: AUG GCG CCG AUA AUG ACG GUC CUU CCU UGA
Protein: Met Ala Pro Ile Met Thr Val Leu Pro StopTranslation (RNA to Protein)
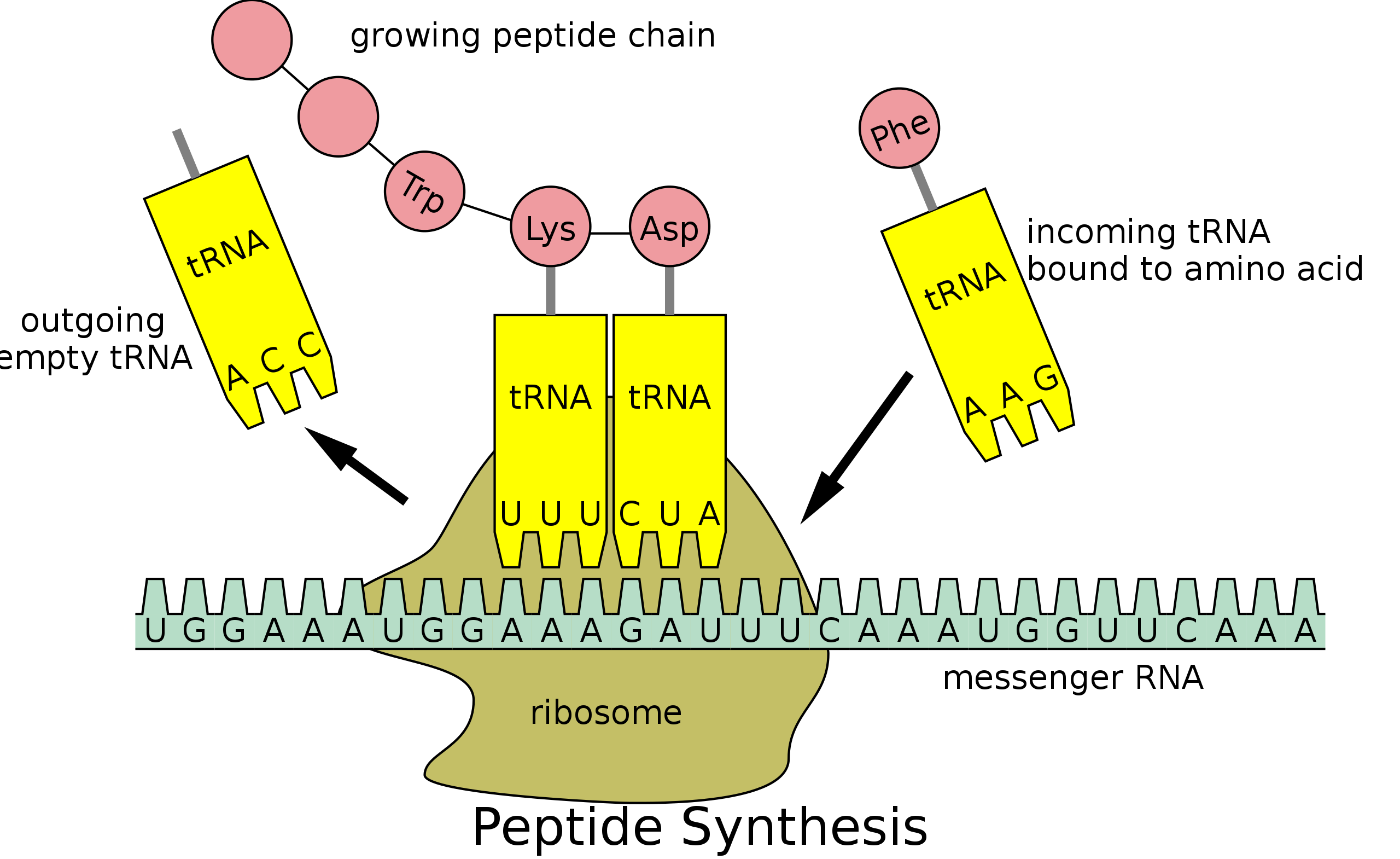
Translation involves the ribosome, a riboprotein complex, which works alongside transfer RNA (tRNA) molecules and various regulatory proteins.
These components ensure that tRNAs are charged with the correct amino acids.
Translation (Basic)
Translation (Intermediate)
Translation (Detailed)
Protein Synthesis
Protein Synthesis
tRNA: 1, 2, 3
Transfer RNA (tRNA)
Transfer RNAs (tRNAs) function as adaptor molecules crucial for protein synthesis.
They are encoded by non-protein-coding genes within the genome.
These genes undergo transcription to produce RNA, which serves as the final functional product.
Transfer RNA (tRNA)
Diversity: Bacteria possess approximately 30 to 45 distinct tRNAs, while eukaryotic organisms can have up to 50 distinct types, with humans having 48.
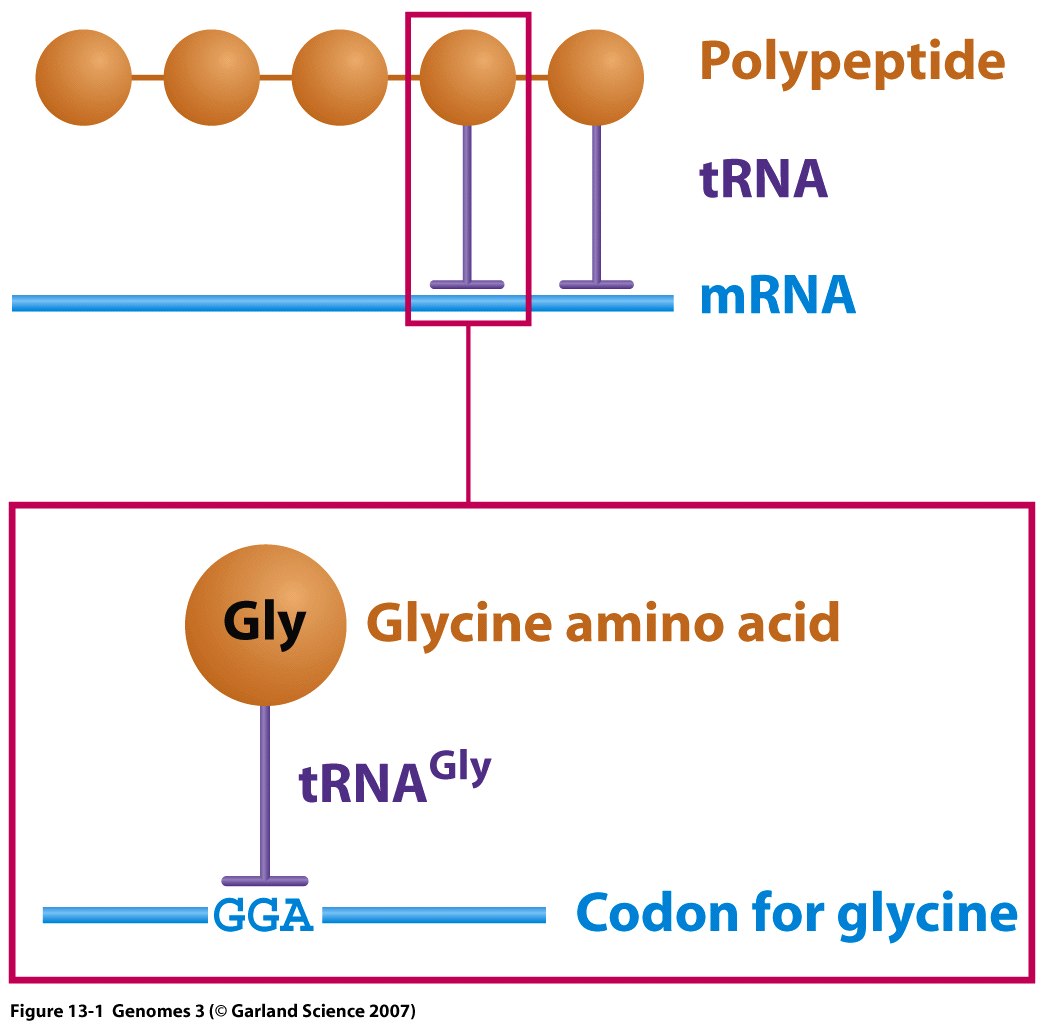
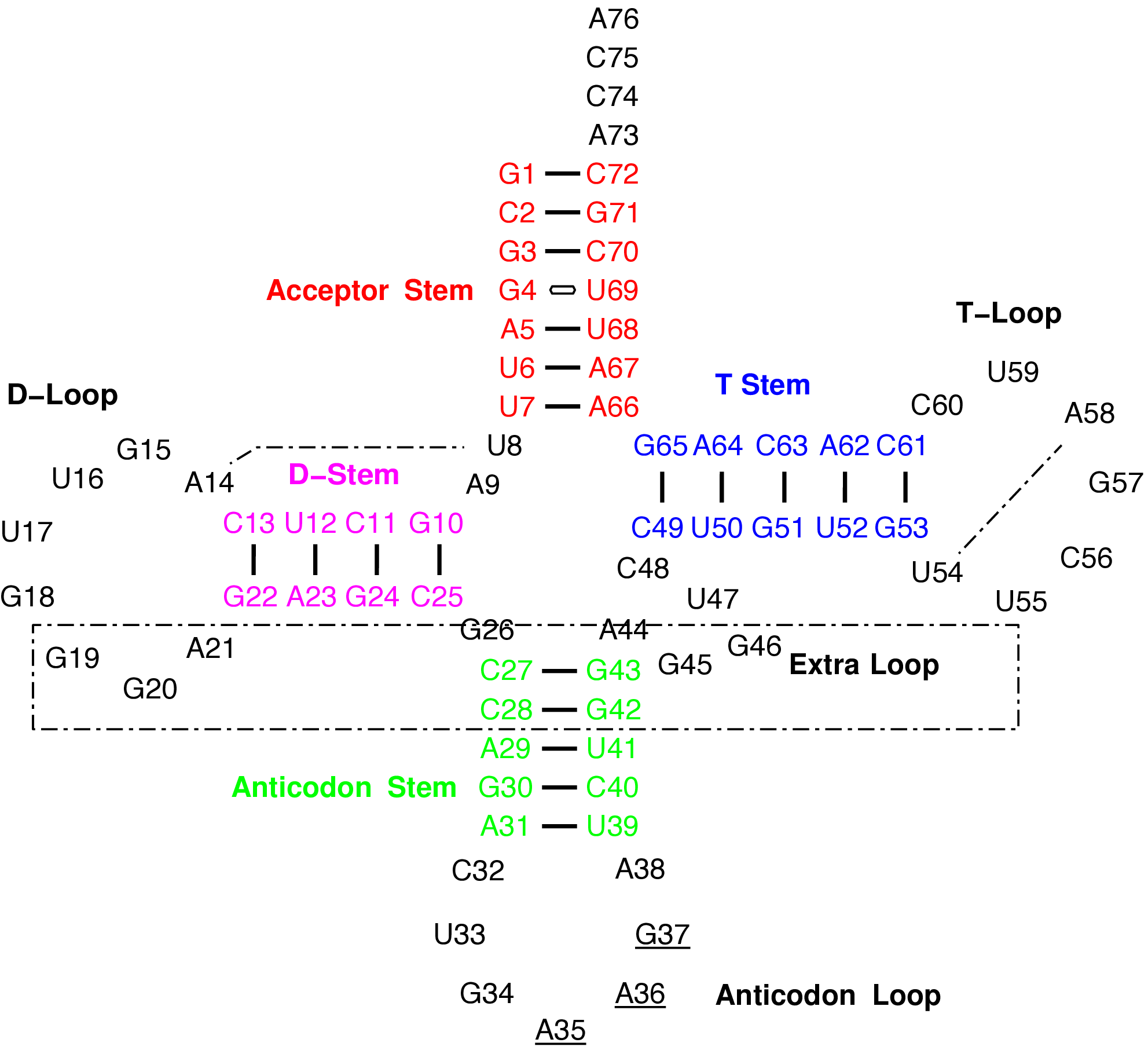
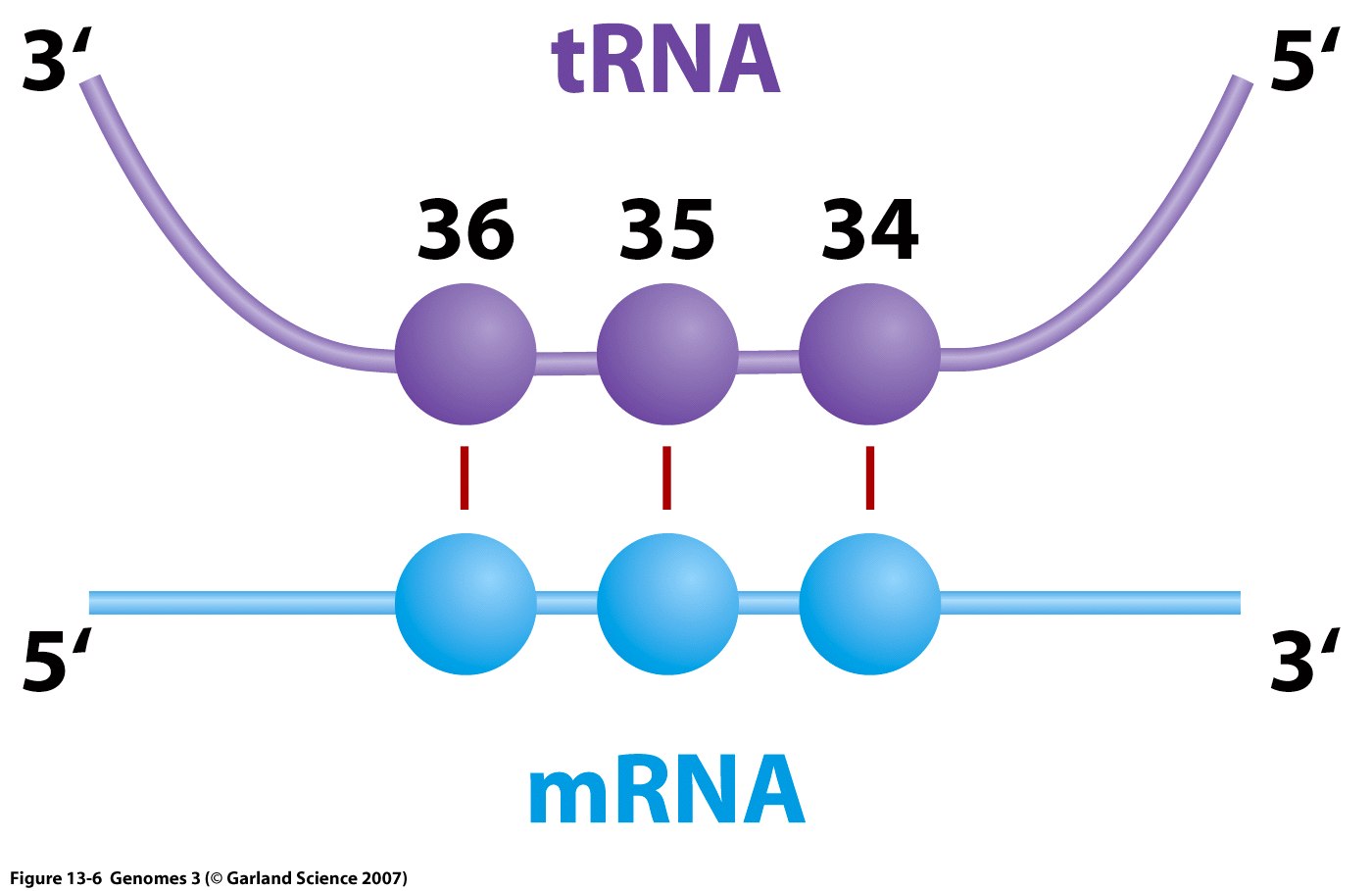
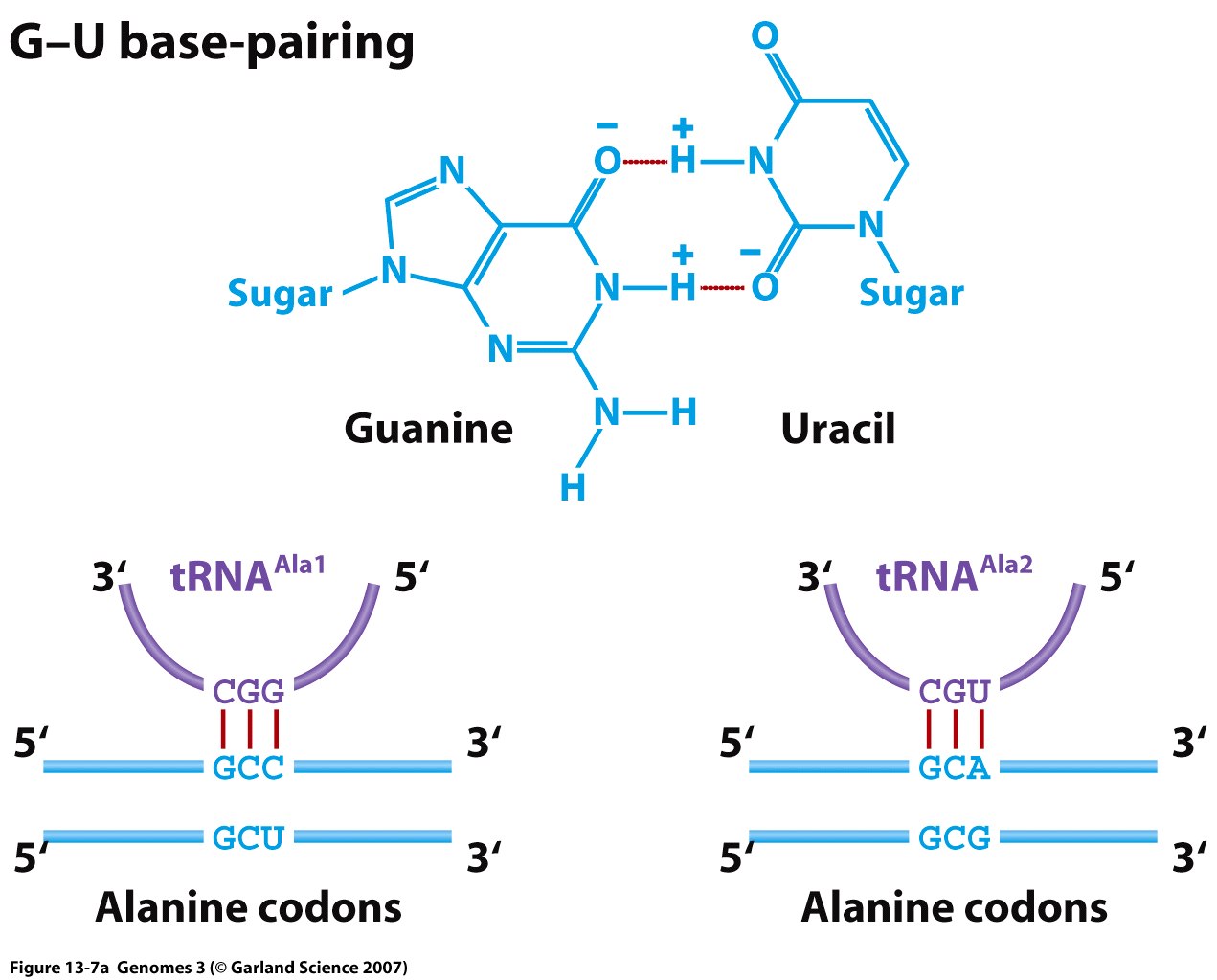
Structure and Function: Each tRNA is covalently bonded to a specific amino acid at one terminus and contains a triplet nucleotide sequence, termed the anti-codon, at the opposite terminus. This sequence is complementary to the mRNA codon.
Transfer RNA (tRNA)
Notation: tRNAPhe denotes a tRNA specifically charged with phenylalanine, one of the 20 standard amino acids.
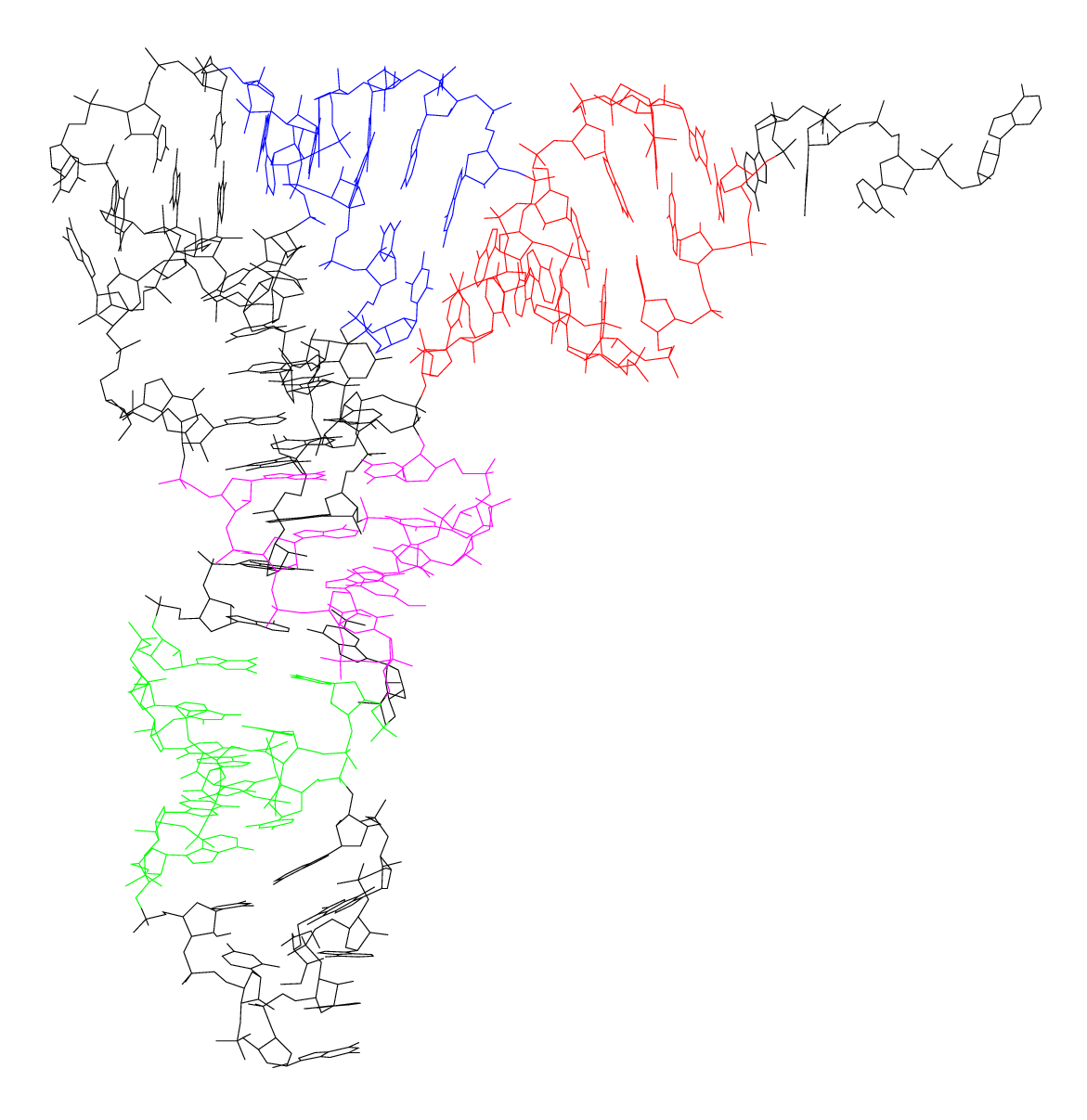
Conformation: Typically, tRNAs are 70 to 90 nucleotides in length, adopting a conserved cloverleaf secondary structure. This common structural motif was illustrated on the preceding slide.
Transfer RNA (tRNA)
- To facilitate protein synthesis, it is essential that all tRNAs exhibit a uniform structure, enabling their efficient interaction with the ribosome.
Transfer RNA (tRNA)
Aminoacyl-tRNA synthetases are enzymes that catalyze the attachment of specific amino acids to their corresponding tRNAs. Typically, organisms possess 20 distinct aminoacyl-tRNA synthetases, each dedicated to linking a particular amino acid to all isoaccepting tRNAs—that is, different tRNAs that are charged with the same type of amino acid.
Each tRNA possesses distinct characteristics that ensure its specific aminoacylation with the correct amino acid.
Protein Synthesis
Protein Synthesis
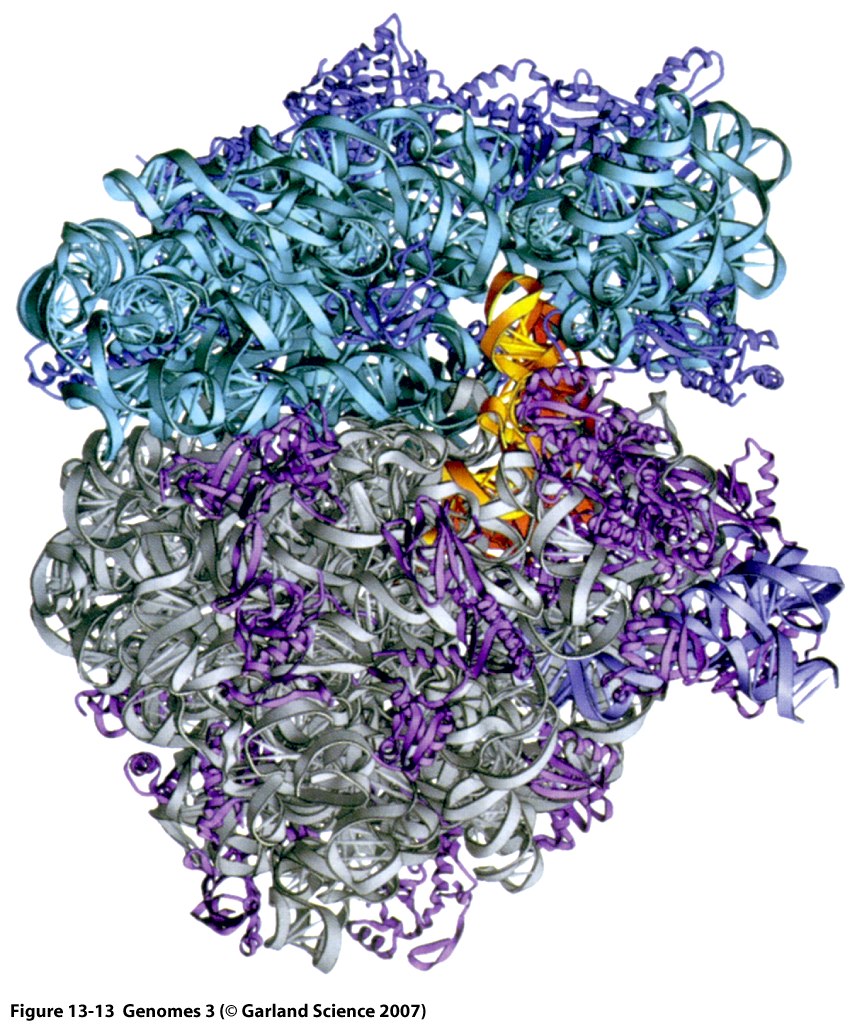
Ribosomes: Key Players in Translation
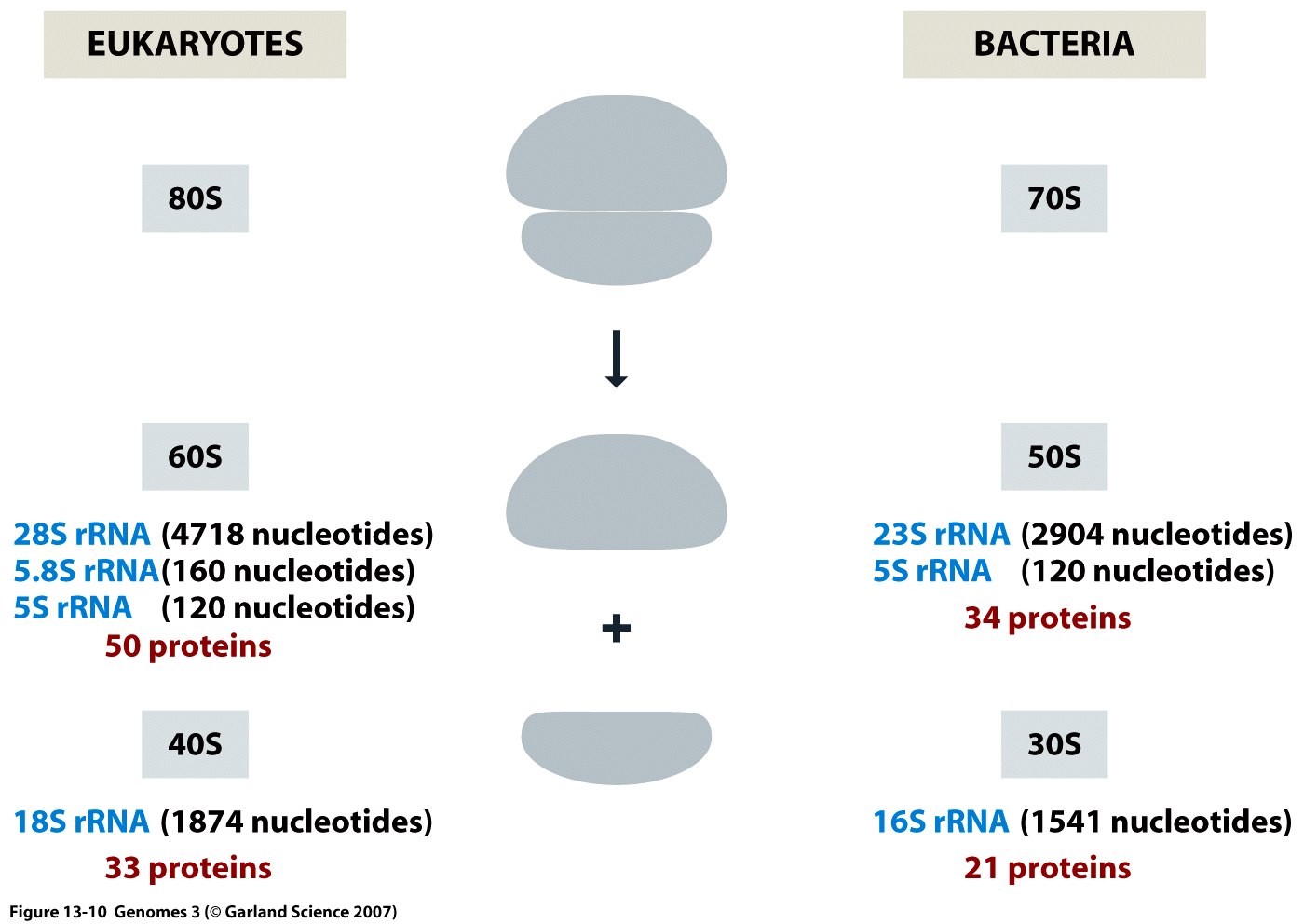
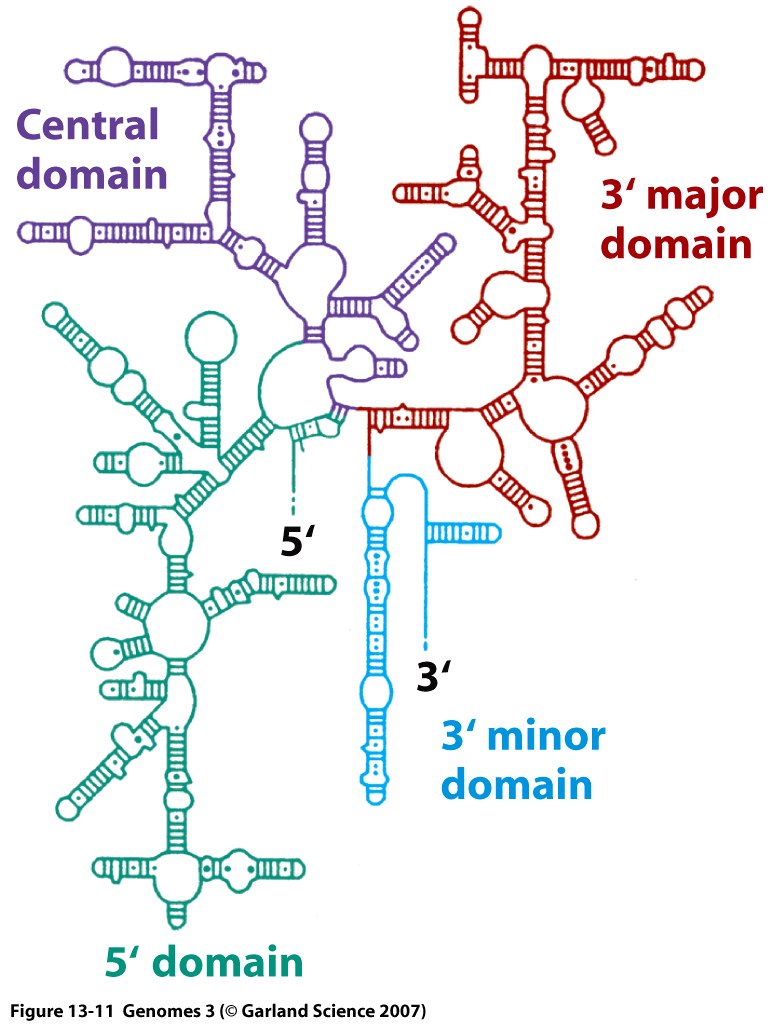
Ribosomes are complex macromolecular structures comprising 3 to 4 RNA molecules and 55 to 83 proteins.
In bacterial cells, ribosome numbers reach approximately 20,000, with eukaryotic cells hosting even more.
Ribosomes facilitate protein synthesis by precisely aligning messenger RNAs (mRNAs), transfer RNAs (tRNAs), and requisite protein factors.
They also play a catalytic role in several biochemical reactions integral to protein synthesis.
Ribosomes
Ribosomes
Ribosomes
DNA-RNA-Protein Relationships
DNA: TAC CGC GCC TAT TAC TGC CAG GAA GGA ACT
RNA: AUG GCG CCG AUA AUG ACG GUC CUU CCU UGA
Protein: M A P I M T V L P *
DNA: TAC CGC GCC TAT TAC TGC CAG GAA GGA ACT
RNA: AUG GCG CCG AUA AUG ACG GUC CUU CCU UGA
Protein: Met Ala Pro Ile Met Thr Val Leu Pro StopReading Frame
Protein translation initiates at the start codon ATG (AUG in RNA), establishing the reading frame. It terminates at a stop codon.
Typically, proteins begin with methionine; however, alternative start codons such as GUG or UUG may be employed in specific mRNAs. Additionally, post-translational modifications can remove the N-terminal segment of the protein.
Reading Frame
There are three stop codons, which are non-sense codons.
Of the 64 possible codons, 61 are sense codons, encoding 20 amino acids, with one codon specifically serving as the start codon, coding for methionine.
The genetic code is described as degenerate because multiple codons can encode the same amino acid. Consequently, a single amino acid sequence may be derived from multiple distinct DNA sequences, ensuring a unique translation.
Worked Example
Summary
- Genetic sequences are organized into triplets known as codons.
- The start codon is AUG, encoding the amino acid methionine (Met).
- There are three stop codons that terminate the polypeptide chain, preventing further amino acid addition.
Summary
- Around 30 to 50 distinct transfer RNAs (tRNAs) exist, each linked to a specific amino acid corresponding to its anticodon sequence. These tRNAs, as nucleic acids, adhere to standard base-pairing rules during codon-anticodon recognition.
Summary
- An Open Reading Frame (ORF) is a sequence of codons beginning with a start codon (AUG) and concluding with a stop codon, defining a potential protein-coding region.
- Given that the genetic code consists of nucleotide triplets, each DNA strand offers three potential reading frames, depending on whether the start codon is positioned at \(i \bmod 3 = 0, 1,\) or \(2\).
- Considering the anti-parallel nature of the two complementary DNA strands, this results in a total of six possible reading frames for translation.
Fun
Fun (continued)
Video playlists by the Amoeba Sisters, “on a mission to demystify science with humor and relevance.”
- Cell Structure and Function (5 videos)
- Biomolecules and Biochemistry (5 videos)
- DNA, RNA, and Protein Synthesis (5 videos)
- DNA Replication, Cell Cycle, and Cell Division (5 videos)
- Viruses, Bacteria, and Cancer (3 videos)
Genome
Genome Sizes
| Species | Size |
|---|---|
| Potato spindle tuber viroid (PSTVd) | 360 |
| Obelisk | 1,000 |
| Human immunodeficiency virus (HIV) | 9,700 |
| SARS-CoV-d (COVID-19) | 29,000 |
| Bacteriophage lambda (\(\lambda\)) | 48,500 |
| Mycoplasma genitalium (bacterium) | 580,000 |
| Escherichia coli (bacterium) | 4,600,000 |
| Ramazzottius varieornatus (tardigrade) | 55,800,000 |
| Drosophila melanogaster (fruit fly) | 120,000,000 |
| Homo sapiens (human) | 3,000 000,000 |
| Bufo bufo (common toad) | 6,900,000,000 |
| Podisma pedestris (mountain grasshopper) | 17,000,000,000 |
| Lilium longiflorum (easter lily) | 90,000,000,000 |
| Necturus lewisi (a salamander) | 118,000,000,000 |
| Amoeba dubia (amoeba) | 670,000,000,000 |
Genome sizes
- Haemophilus influenzae (bacterium), dna = 1.8 Mbp
- Escherichia coli (bacterium), dna = 4.6 Mbp
- Saccharomyces cerevisiae (yeast), dna = 12 Mbp
- Caenorhabditis elegans (worm), dna = 97 Mbp
- Arabidopsis thaliana (flowering plant), dna = 115 Mbp
- Drosophila melanogaster (fruit fly), dna = 137 Mbp
- Smallest Human chromosome (Y), dna = 50 Mbp
- Largest Human chromosome (1), dna = 250 Mbp
- Whole Human genome, dna = 3 Gbp
- Mus musculus (mouse), dna = 3 Gbp.
DNA is organized into chromosomes
The self-replicating genetic structures of cells containing the cellular DNA that bears in its nucleotide sequence the linear array of genes. In prokaryotes, chromosomal DNA is circular, and the entire genome is carried on one chromosome. Eukaryotic genomes consist of a number of chromosomes whose DNA is associated with different kinds of proteins.
Genome of Multicellular Animals
The human genome consists of two primary components: nuclear and mitochondrial genome.
Genome of Multicellular Animals
- Nuclear Genome: The nuclear genome comprises 23 chromosome pairs, amounting to 24 unique linear DNA molecules: 22 autosomes and the X and Y sex chromosomes. Chromosomal lengths range from about 50 million nucleotides for the shortest to over 205 million for the longest. Cumulatively, the nuclear genome encompasses approximately 3.2 billion nucleotides and encodes approximately 20,000 protein-coding genes.
Genome of Multicellular Animals
- Mitochondrial Genome: The mitochondrial genome comprises a single circular DNA molecule, 16,569 nucleotides in length, present in multiple copies within the mitochondria. It encodes 37 genes essential for protein synthesis.
Genomic Composition Across Cells
- The adult human body comprises approximately \(10^{13}\) cells, each possessing an identical genomic sequence.
Human Chromosomal Composition
- Human somatic cells are diploid, containing two sets of the 22 autosomes and two sex chromosomes: XX in females, XY in males.
- Somatic cells, synonymous with diploid cells, contrast with gametes.
- Gametes are haploid, possessing only one set of the 22 autosomes and a single sex chromosome.
Genes
What are the genes?
The fundamental physical and functional unit of heredity. A gene is an ordered sequence of nucleotides located in a particular position on a particular chromosome that encodes a specific functional product (i.e., a protein or RNA molecule).
- Can be several thousands nt (nucleotides) long.
- Occurs on either strand, not often but sometimes overlapping.
Genome Organisation
In higher organisms, protein-coding genes are composed of subsegments known as exons, which are interspersed with intervening sequences termed introns.
Genomic Organization
Contrary to common perception, genomes are not densely packed with genes.
Genomic Organization
In the human genome, the structure is as follows:
- Approximately 60% consists of repetitive sequences:
- One-third of these are satellite DNAs, characterized by low complexity, short length, and high repetition.
- The remaining two-thirds are composed of complex repeats, including transposable elements and similar structures.
Genomic Organization (continued)
- Unique sequences contribute to a smaller portion:
- Only 1.2% are protein-coding regions.
- Introns account for around 20% of the genome.
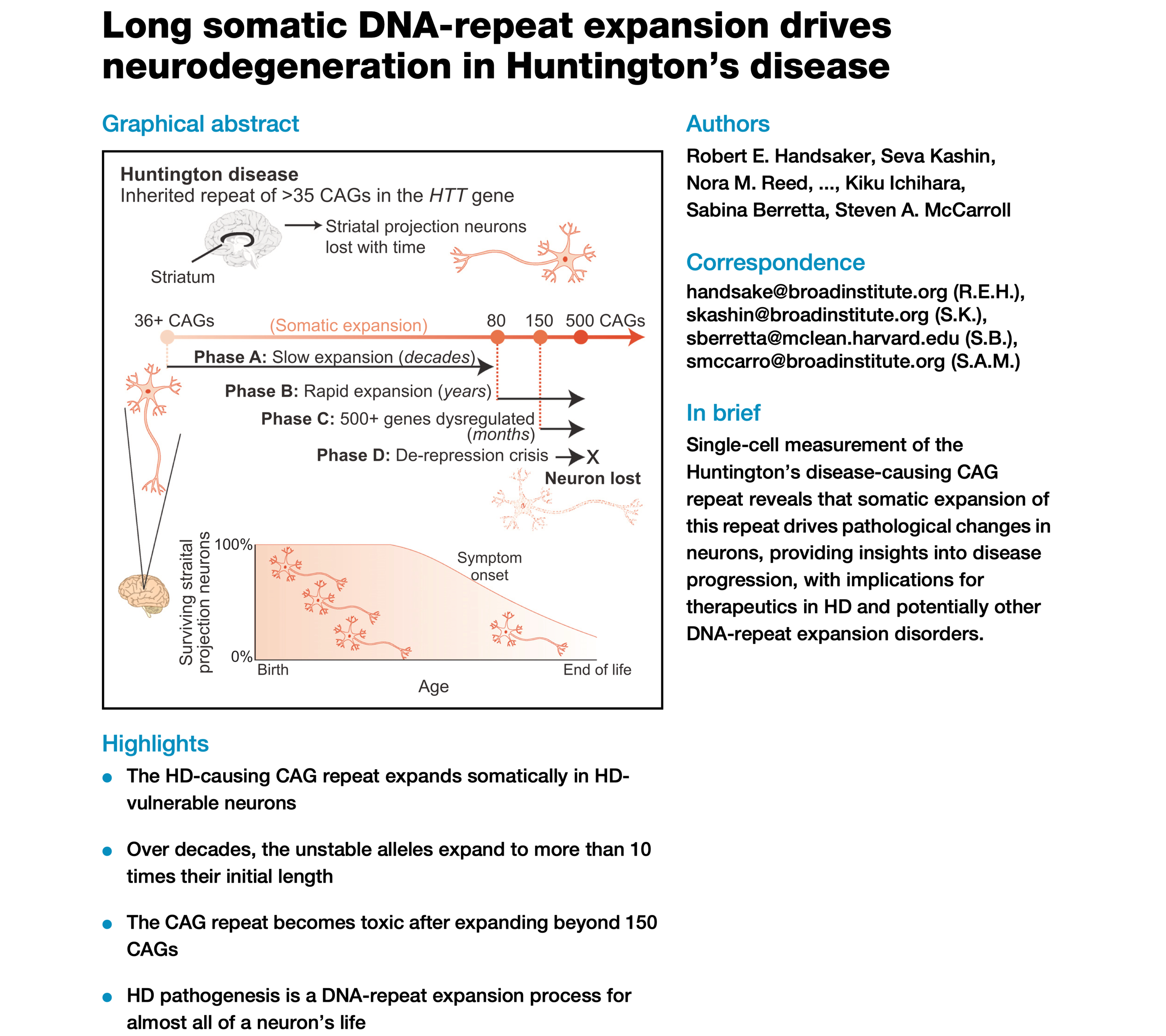
DNA-Repeat Expansion & Diseases
Genome Organisation
- “About one-half of the platypus genome consists of interspersed repeats derived from transposable elements.”
Platypus
The platypus is a unique, semi-aquatic mammal native to eastern Australia, including Tasmania. It is notable for its unusual combination of features: a duck-bill, webbed feet, and a beaver-like tail. Unlike most mammals, it lays eggs and has venomous spurs on the males’ hind legs. The platypus is one of the few monotremes, a primitive group of egg-laying mammals, and it uses electroreception to locate prey underwater.
Bioinformaticist’s Perspective
- DNA Sequencing (traditional or high-throughput)
- Gene Finding (stochastic grammatical models)
- Identifying Signals (pattern discovery, now deep learning)
Bioinformaticist’s Perspective
Repetitive sequences pose significant challenges to algorithms employed in sequence assembly due to their complex structures and redundancy.
The association of repetitive sequences with various diseases underscores their detection as a critical area of research in bioinformatics.
Proteome
Proteome
- The proteome encompasses the complete set of proteins expressed by an organism at a specific time, while proteomics investigates the interactions and functions of these proteins.
Proteome
- Analogous to the transcriptome, the proteome is inherently dynamic, reflecting the organism’s physiological state and environmental interactions.
Proteome
- Proteins serve as critical cellular components, not only forming structural elements but also catalyzing the majority of biochemical reactions.
Proteome
- As Brown (2006) notes, understanding how a genome defines a cell’s biochemical capabilities remains a central challenge in contemporary biology.
Proteome
- There is a paradigm shift from traditional hypothesis-driven, reductionist methodologies to holistic, data-driven, systems-based approaches.
Networks
Interaction Networks
- Protein-Protein interactions (PPI)
- Protein-DNA interactions
- Genetic interactions
- Metabolic networks
- Signaling network
- Transcription/regulatory network
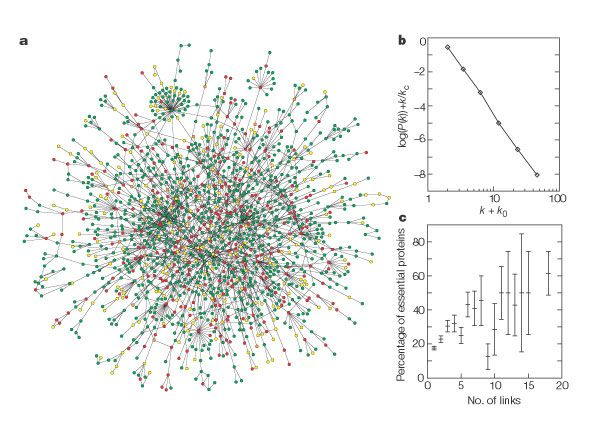
Yeast Proteome
Metabolic network
Resources
Prologue
Summary
We investigated the translation phase of the central dogma, emphasizing its reliance on the genetic code, codons, and tRNA-facilitated amino acid transport.
We analyzed genome organization, focusing on repetitive DNA sequences and their implications for assembly algorithms and disease pathogenesis.
In conclusion, we delineated proteomic principles and the complex networks of biological interactions, highlighting the intricate nature of gene expression and regulation.
Central Dogma (Futuristic Animation)
Next Lecture
- Essential Bioinformatics
References
Marcel Turcotte
School of Electrical Engineering and Computer Science (EECS)
University of Ottawa