import requestsEssential Bioinformatics - Get Transcript
CSI 5180 - Machine Learning for Bioinformatics
Learning Objectives
- Develop a basic understanding of Jupyter Notebooks and their applications in computational research.
- Implement code execution within Jupyter Notebooks by utilizing Google Colab as a cloud-based platform.
- Adapt existing Jupyter Notebooks by making minor modifications and subsequently executing the revised code cells within the Google Colab environment.
Quick Start Guide
Interactive Exploration Without Installation
This notebook is designed for seamless online use, eliminating the need for local installation. It is important to note that the code detailed in this notebook requires the requests library, which is conveniently pre-installed on Google Colab. To execute the code on Colab, a Google account is necessary.
If you are accessing this document on the course website, you are viewing an HTML-rendered version of a Jupyter Notebook (not the Jupyter Notebook itself). To download the Jupyter Notebook directly, navigate to the table of contents, usually located in the upper right corner of your browser window, or download it directly via this link: 01_get_transcript.ipynb.
View the Notebook Without Code Execution
For a static view of the notebook without executing any code, you can access it via nbviewer:
Concepts
A Jupyter notebook serves as an interactive computational platform that integrates code execution, textual content, and multimedia into a unified document. The primary components of a Jupyter notebook are as follows:
Cells
- Code Cells: These cells are designed to hold executable code. While Python is the most commonly used language, other languages such as R, Julia, and Scala are also supported. The code within these cells is executed via the kernel, and the resulting output is displayed immediately below the cell.
- Markdown Cells: These cells are used for text formatted in Markdown. They facilitate the inclusion of explanations, headers, bullet points, hyperlinks, images, and various other text formats within the notebook.
- Raw Cells: These cells contain text that is neither executed nor rendered. They are particularly useful for embedding code snippets or unformatted text that should remain unchanged.
Execution
- Standard Output: The results of code execution, encompassing text, tables, and plots, are presented directly below the corresponding code cell.
- Rich Media Output: Jupyter notebooks are capable of delivering rich media output, including HTML, images, videos, LaTeX, and interactive widgets, such as those provided by Plotly and Bokeh.
Example
To ensure this example is self-contained, we will utilize the Ensembl REST API.
Ensembl serves as a genome browser for vertebrate genomes, facilitating research in comparative genomics, evolutionary biology, sequence variation, and transcriptional regulation. It provides gene annotations, computes multiple alignments, predicts regulatory functions, and aggregates disease-related data. Ensembl’s suite of tools includes BLAST, BLAT, BioMart, and the Variant Effect Predictor (VEP), applicable to all supported species.
We will employ the Python requests library to interact with the API. Let’s begin by importing this library.
We will define a function that leverages the Ensembl REST API to retrieve the sequence of a specified transcript.
def get_transcript_info(transcript_id):
"""Fetches information about a given transcript from the Ensembl REST API."""
server = "https://rest.ensembl.org"
endpoint = f"/lookup/id/{transcript_id}"
headers = {"Content-Type": "application/json"}
response = requests.get(server + endpoint, headers=headers)
if not response.ok:
response.raise_for_status()
return response.json()The newly defined function is now being executed.
# A variable to store the ID of the transcript of interest
transcript_id = "ENST00000288602"
# Fetch and print transcript information
transcript_info = get_transcript_info(transcript_id)
print(f"Transcript ID: {transcript_info['id']}")
print(f"Gene Name: {transcript_info.get('display_name', 'N/A')}")
print(f"Transcript Name: {transcript_info.get('id', 'N/A')}")
print(f"Biotype: {transcript_info.get('biotype', 'N/A')}")
print(f"Species: {transcript_info.get('species', 'N/A')}")
print()Transcript ID: ENST00000288602
Gene Name: BRAF-201
Transcript Name: ENST00000288602
Biotype: protein_coding
Species: homo_sapiens
The results of the execution have been incorporated into the notebook.
def get_cdna_sequence(transcript_id):
"""Fetches the cDNA sequence of a given transcript from the Ensembl REST API."""
server = "https://rest.ensembl.org"
endpoint = f"/sequence/id/{transcript_id}"
headers = {"Content-Type": "application/json"}
response = requests.get(server + endpoint, headers=headers, params={"type": "cdna"})
if not response.ok:
response.raise_for_status()
data = response.json()
return data['seq']Executing the function get_cdna_sequence. Libraries (e.g. requests), functions (e.g. get_cdna_sequence), and variables (e.g. transcript_id) that were imported or defined in previously executed cells are readily accessible for the current execution cell.
# Fetch cDNA sequence and find start codons
cdna_sequence = get_cdna_sequence(transcript_id)
print(cdna_sequence)CCGCTCGGGCCCCGGCTCTCGGTTATAAGATGGCGGCGCTGAGCGGTGGCGGTGGTGGCGGCGCGGAGCCGGGCCAGGCTCTGTTCAACGGGGACATGGAGCCCGAGGCCGGCGCCGGCGCCGGCGCCGCGGCCTCTTCGGCTGCGGACCCTGCCATTCCGGAGGAGGTGTGGAATATCAAACAAATGATTAAGTTGACACAGGAACATATAGAGGCCCTATTGGACAAATTTGGTGGGGAGCATAATCCACCATCAATATATCTGGAGGCCTATGAAGAATACACCAGCAAGCTAGATGCACTCCAACAAAGAGAACAACAGTTATTGGAATCTCTGGGGAACGGAACTGATTTTTCTGTTTCTAGCTCTGCATCAATGGATACCGTTACATCTTCTTCCTCTTCTAGCCTTTCAGTGCTACCTTCATCTCTTTCAGTTTTTCAAAATCCCACAGATGTGGCACGGAGCAACCCCAAGTCACCACAAAAACCTATCGTTAGAGTCTTCCTGCCCAACAAACAGAGGACAGTGGTACCTGCAAGGTGTGGAGTTACAGTCCGAGACAGTCTAAAGAAAGCACTGATGATGAGAGGTCTAATCCCAGAGTGCTGTGCTGTTTACAGAATTCAGGATGGAGAGAAGAAACCAATTGGTTGGGACACTGATATTTCCTGGCTTACTGGAGAAGAATTGCATGTGGAAGTGTTGGAGAATGTTCCACTTACAACACACAACTTTGTACGAAAAACGTTTTTCACCTTAGCATTTTGTGACTTTTGTCGAAAGCTGCTTTTCCAGGGTTTCCGCTGTCAAACATGTGGTTATAAATTTCACCAGCGTTGTAGTACAGAAGTTCCACTGATGTGTGTTAATTATGACCAACTTGATTTGCTGTTTGTCTCCAAGTTCTTTGAACACCACCCAATACCACAGGAAGAGGCGTCCTTAGCAGAGACTGCCCTAACATCTGGATCATCCCCTTCCGCACCCGCCTCGGACTCTATTGGGCCCCAAATTCTCACCAGTCCGTCTCCTTCAAAATCCATTCCAATTCCACAGCCCTTCCGACCAGCAGATGAAGATCATCGAAATCAATTTGGGCAACGAGACCGATCCTCATCAGCTCCCAATGTGCATATAAACACAATAGAACCTGTCAATATTGATGACTTGATTAGAGACCAAGGATTTCGTGGTGATGGAGCCCCTTTGAACCAGCTGATGCGCTGTCTTCGGAAATACCAATCCCGGACTCCCAGTCCCCTCCTACATTCTGTCCCCAGTGAAATAGTGTTTGATTTTGAGCCTGGCCCAGTGTTCAGAGGATCAACCACAGGTTTGTCTGCTACCCCCCCTGCCTCATTACCTGGCTCACTAACTAACGTGAAAGCCTTACAGAAATCTCCAGGACCTCAGCGAGAAAGGAAGTCATCTTCATCCTCAGAAGACAGGAATCGAATGAAAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGACAAAGAATTGGATCTGGATCATTTGGAACAGTCTACAAGGGAAAGTGGCATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAGCACCTACACCTCAGCAGTTACAAGCCTTCAAAAATGAAGTAGGAGTACTCAGGAAAACACGACATGTGAATATCCTACTCTTCATGGGCTATTCCACAAAGCCACAACTGGCTATTGTTACCCAGTGGTGTGAGGGCTCCAGCTTGTATCACCATCTCCATATCATTGAGACCAAATTTGAGATGATCAAACTTATAGATATTGCACGACAGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAATAATATATTTCTTCATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATCAGTTTGAACAGTTGTCTGGATCCATTTTGTGGATGGCACCAGAAGTCATCAGAATGCAAGATAAAAATCCATACAGCTTTCAGTCAGATGTATATGCATTTGGAATTGTTCTGTATGAATTGATGACTGGACAGTTACCTTATTCAAACATCAACAACAGGGACCAGATAATTTTTATGGTGGGACGAGGATACCTGTCTCCAGATCTCAGTAAGGTACGGAGTAACTGTCCAAAAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAAAGAAAAGAGATGAGAGACCACTCTTTCCCCAAATTCTCGCCTCTATTGAGCTGCTGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCATCAGAACCCTCCTTGAATCGGGCTGGTTTCCAAACAGAGGATTTTAGTCTATATGCTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGGGATATGGTGCGTTTCCTGTCCACTGAAACAAATGAGTGAGAGAGTTCAGGAGAGTAGCAACAAAAGGAAAATAAATGAACATATGTTTGCTTATATGTTAAATTGAATAAAATACTCTCTTTTTTTTTAAGGTGAACWe are now focused on identifying all potential start codons.
def find_start_codons(sequence):
"""Finds and returns the positions of all start codons (ATG) in the sequence."""
start_codon = "ATG"
start_codon_positions = []
for i in range(len(sequence) - 2):
if sequence[i:i+3] == start_codon:
start_codon_positions.append(i + 1) # +1 to convert to 1-based indexing
return start_codon_positionsExecuting the function find_start_codons.
# Find start codons
start_codon_positions = find_start_codons(cdna_sequence)
print(f"Start codon positions in the cDNA sequence of {transcript_id}:")
print(start_codon_positions)Start codon positions in the cDNA sequence of ENST00000288602:
[30, 96, 186, 274, 298, 378, 457, 585, 588, 634, 697, 715, 818, 864, 877, 1078, 1132, 1168, 1201, 1224, 1461, 1492, 1510, 1579, 1585, 1599, 1606, 1648, 1678, 1698, 1797, 1839, 1903, 1958, 2007, 2028, 2062, 2068, 2089, 2097, 2151, 2214, 2226, 2254, 2386, 2428, 2456, 2499, 2507, 2519]To illustrate the application of Jupyter Notebooks for data visualization, consider a straightforward example. Notably, Jupyter Notebooks are highly effective tools for conducting exploratory data analysis.
import matplotlib.pyplot as plt
from collections import Counter
def plot_nucleotide_histogram(sequence):
"""Plots a histogram of nucleotide frequencies in the sequence."""
nucleotide_counts = Counter(sequence)
nucleotides = ['A', 'T', 'C', 'G']
counts = [nucleotide_counts[nucleotide] for nucleotide in nucleotides]
plt.bar(nucleotides, counts, color=['blue', 'red', 'green', 'purple'])
plt.title('Nucleotide Frequency Histogram')
plt.xlabel('Nucleotide')
plt.ylabel('Frequency')
plt.show()
# Plot nucleotide frequency histogram
plot_nucleotide_histogram(cdna_sequence)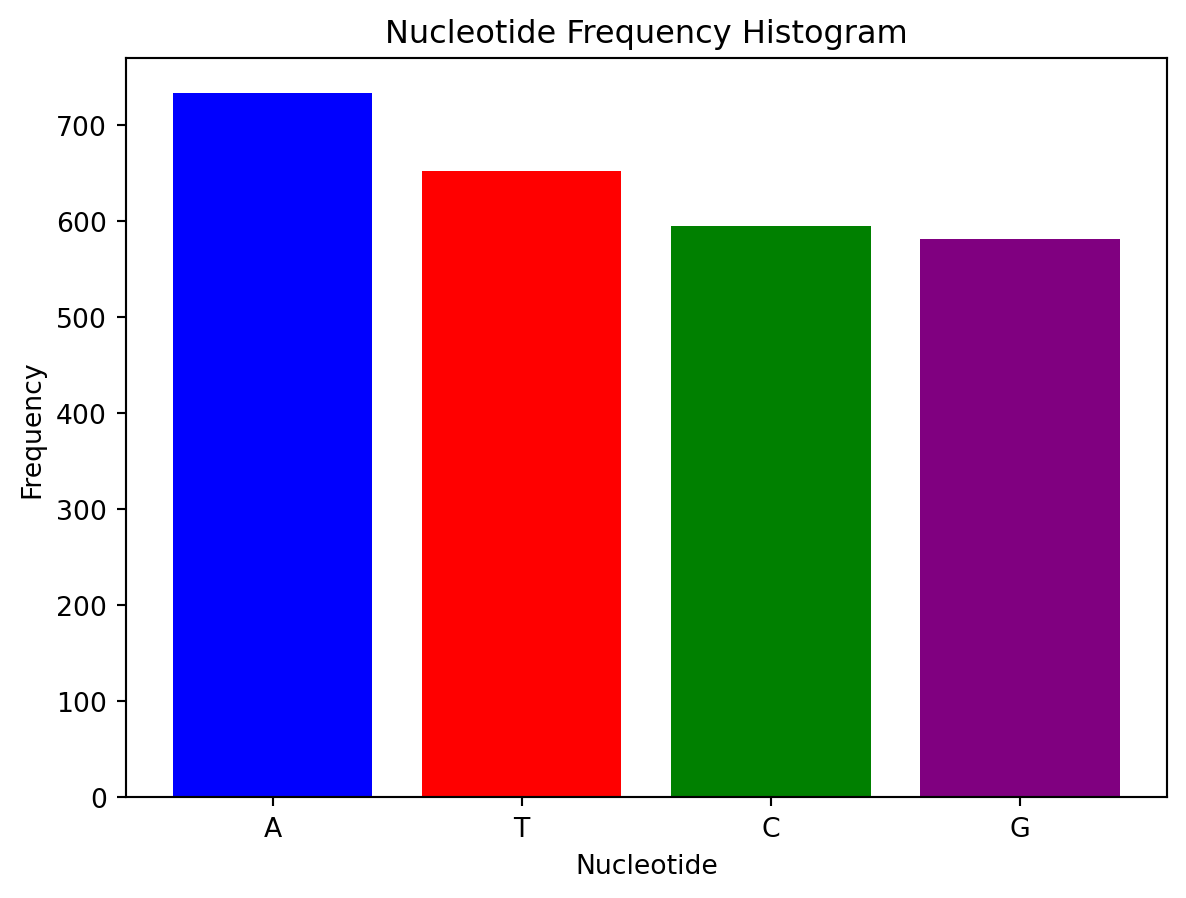
In the previous example, the plot_nucleotide_histogram function is invoked within the same cell where it is defined. This setup results in the redefinition of the function each time it is executed. Furthermore, any modification to the function necessitates re-execution of the cell, which automatically triggers the function call. To enhance clarity and maintainability, I advocate for separating function definition from invocation, as demonstrated earlier. This separation not only improves code readability but also facilitates iterative development and debugging.
Exercise
- Write function that identifies the first open reading frame.