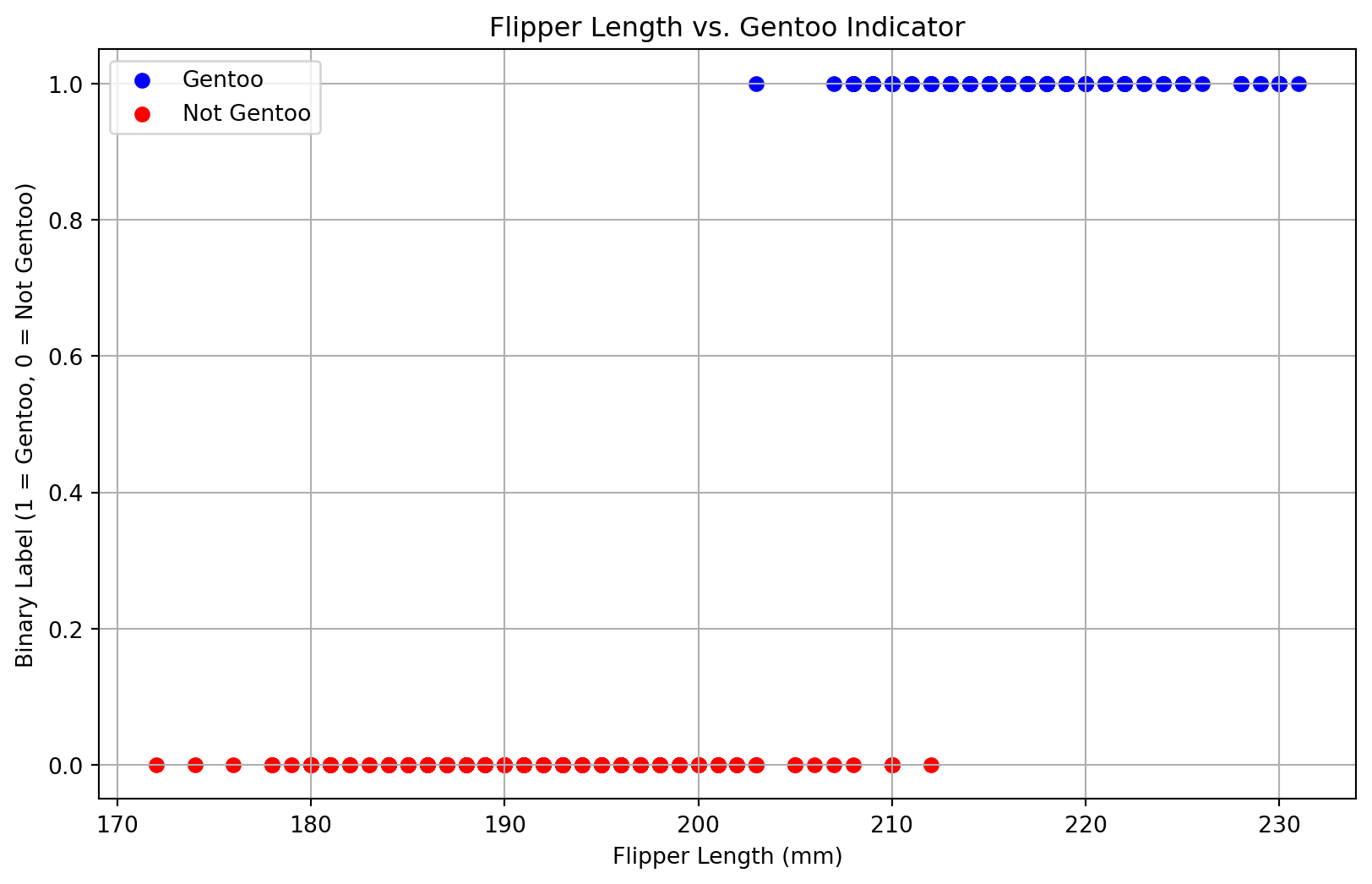
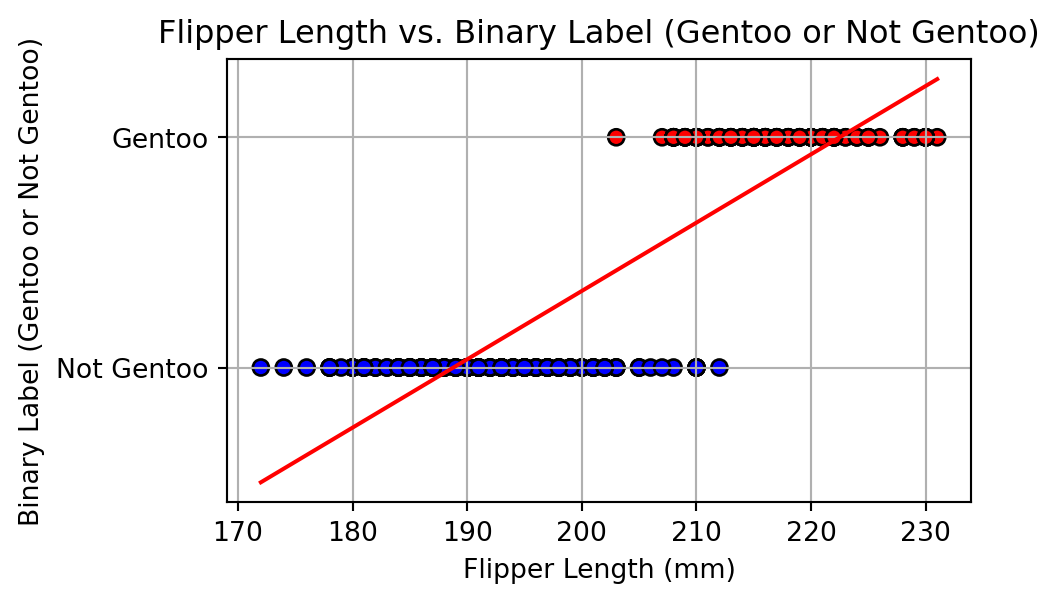
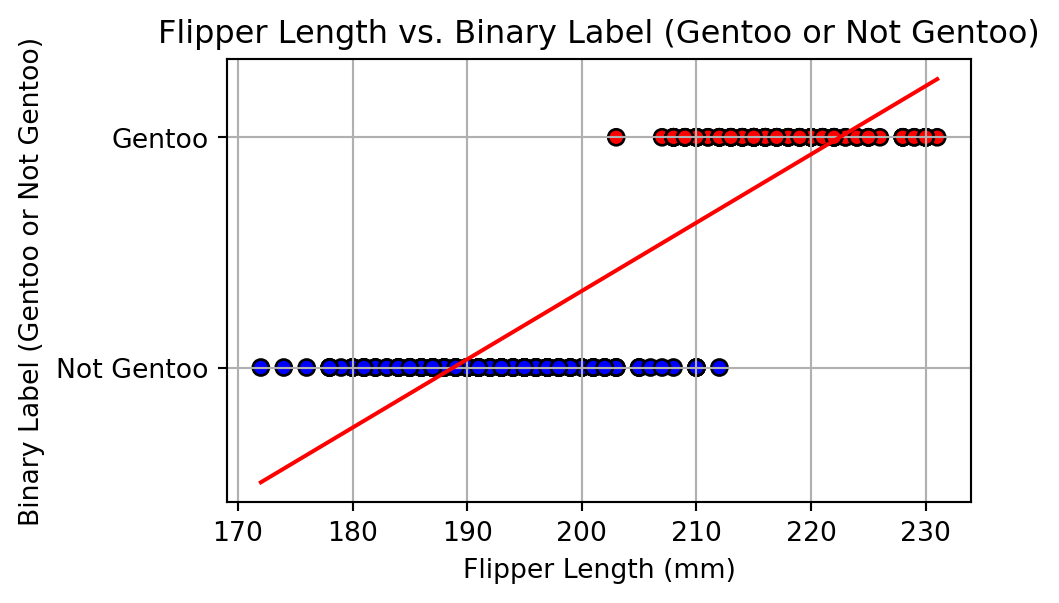
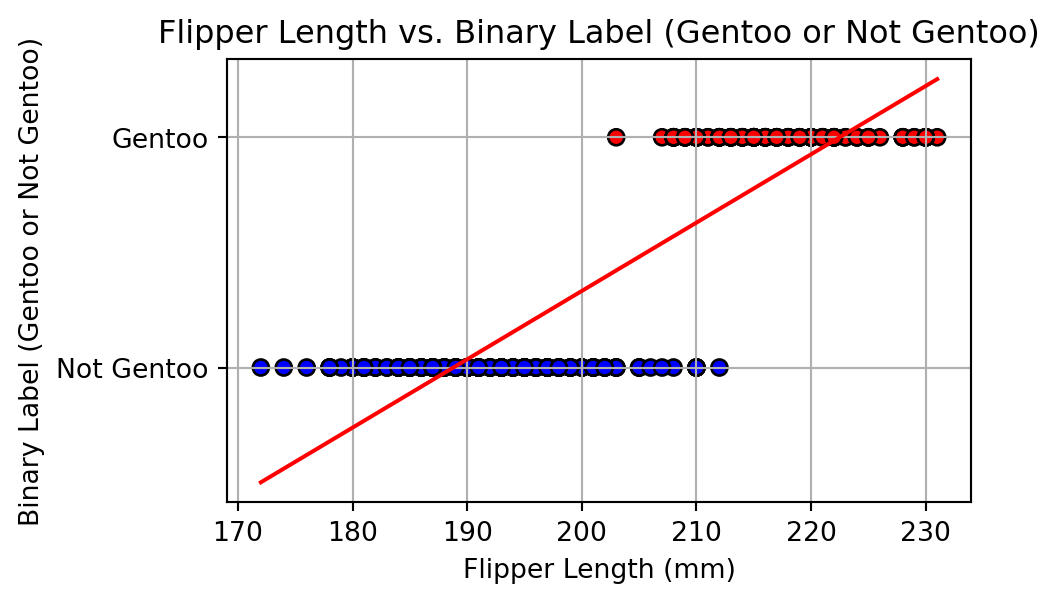
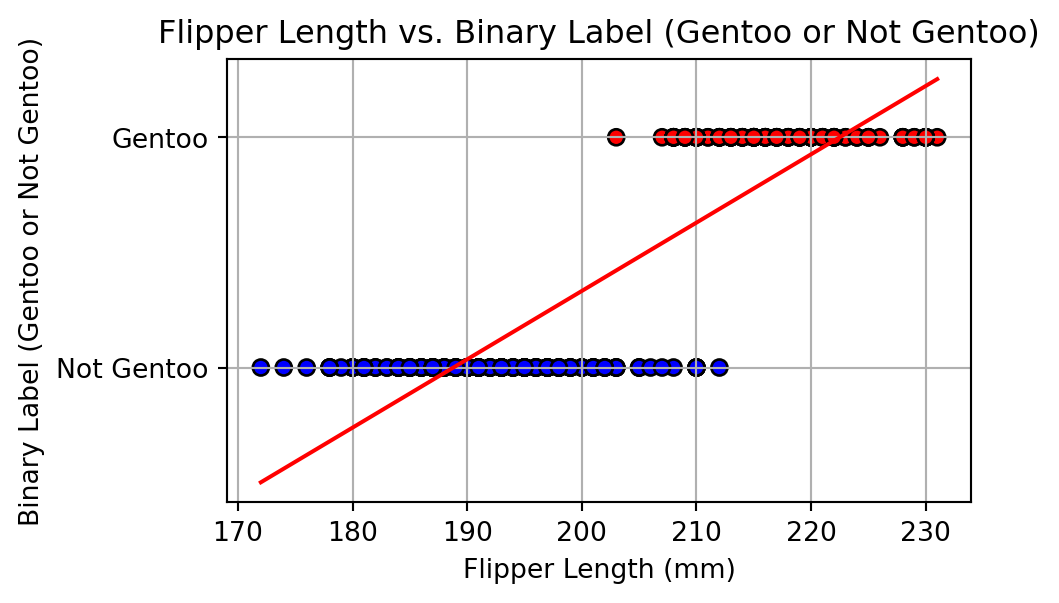
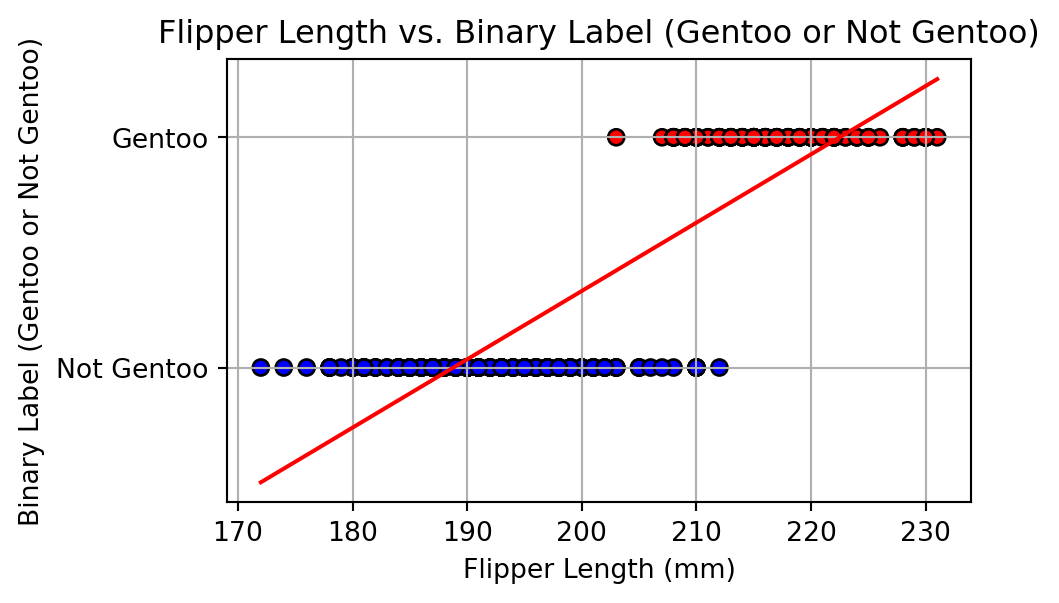
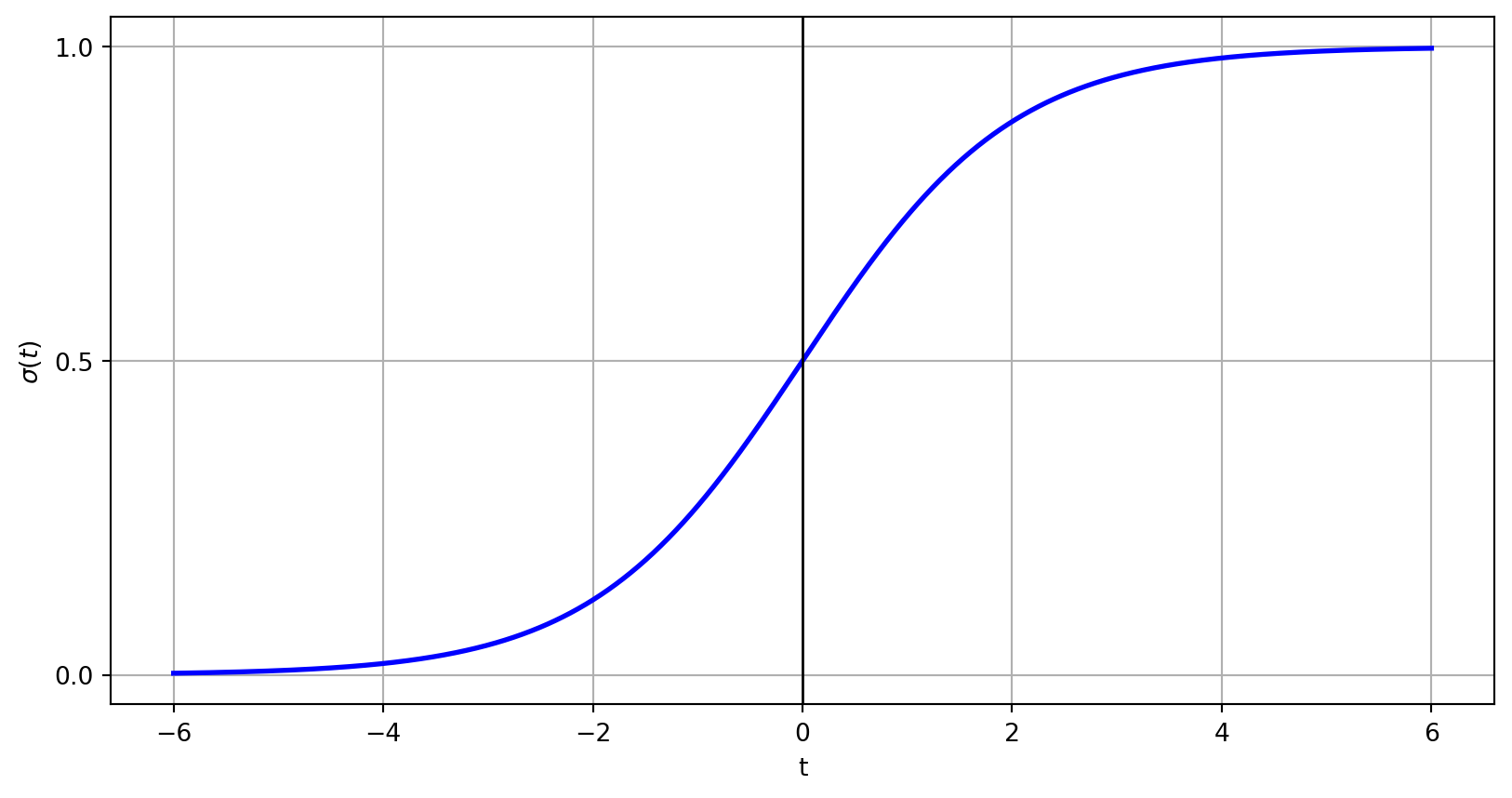
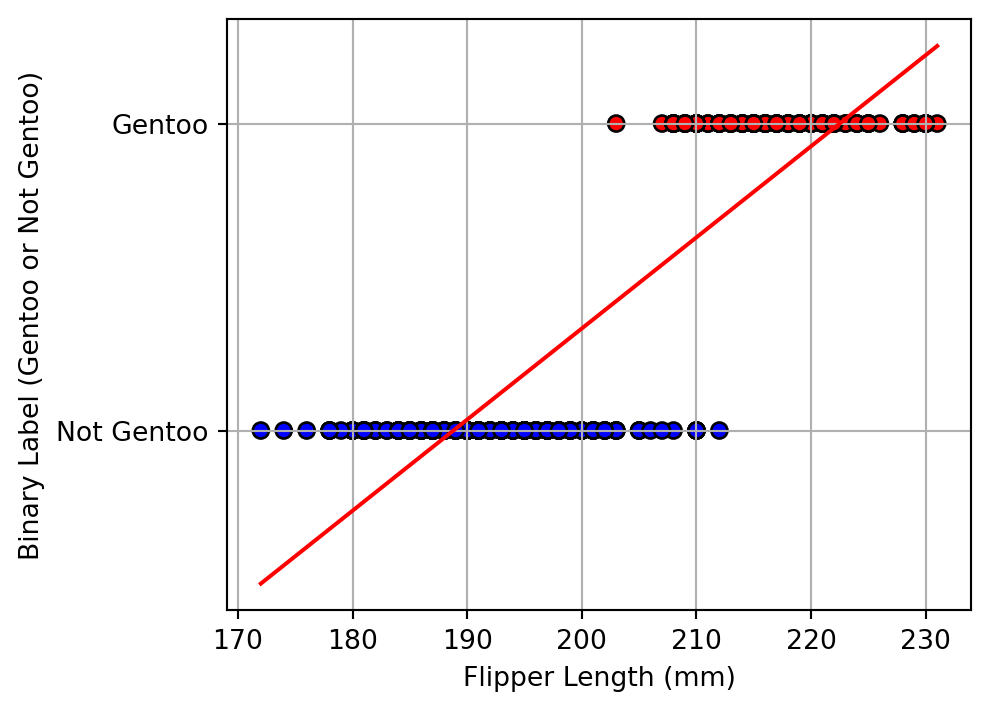
Alharbi, Fadi, and Aleksandar Vakanski. 2023.
“Machine Learning Methods for Cancer Classification Using Gene Expression Data: A Review.” Bioengineering 10 (2): 173.
https://doi.org/10.3390/bioengineering10020173.
Torang, Arezo, Paraag Gupta, and David J. Klinke. 2019.
“An elastic-net logistic regression approach to generate classifiers and gene signatures for types of immune cells and T helper cell subsets.” BMC Bioinformatics 20 (1): 433.
https://doi.org/10.1186/s12859-019-2994-z.
Wu, Qianfan, Adel Boueiz, Alican Bozkurt, Arya Masoomi, Allan Wang, Dawn L DeMeo, Scott T Weiss, and Weiliang Qiu. 2018.
“Deep Learning Methods for Predicting Disease Status Using Genomic Data.” Journal of Biometrics & Biostatistics 9 (5).