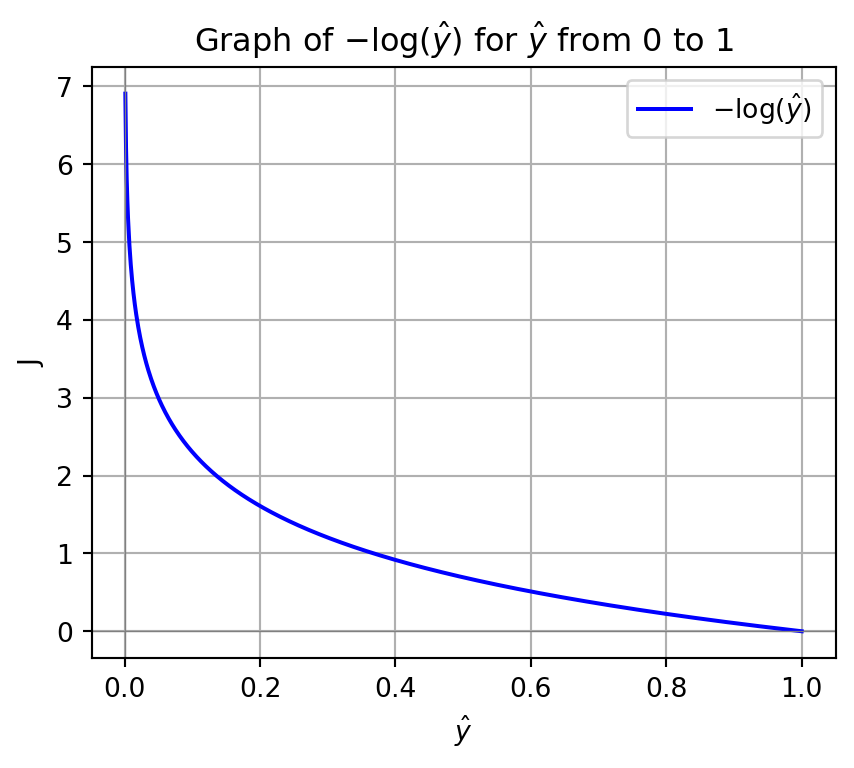
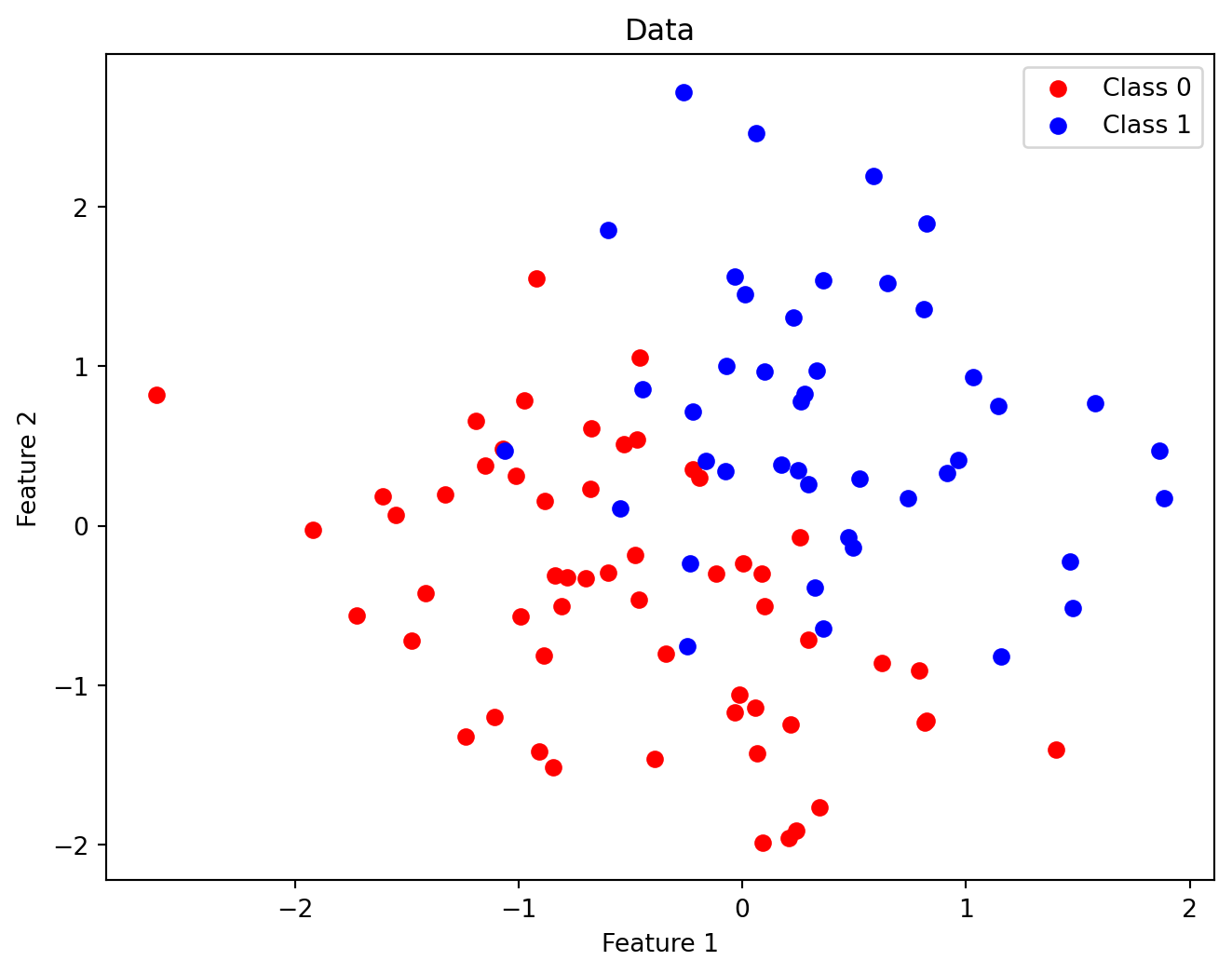
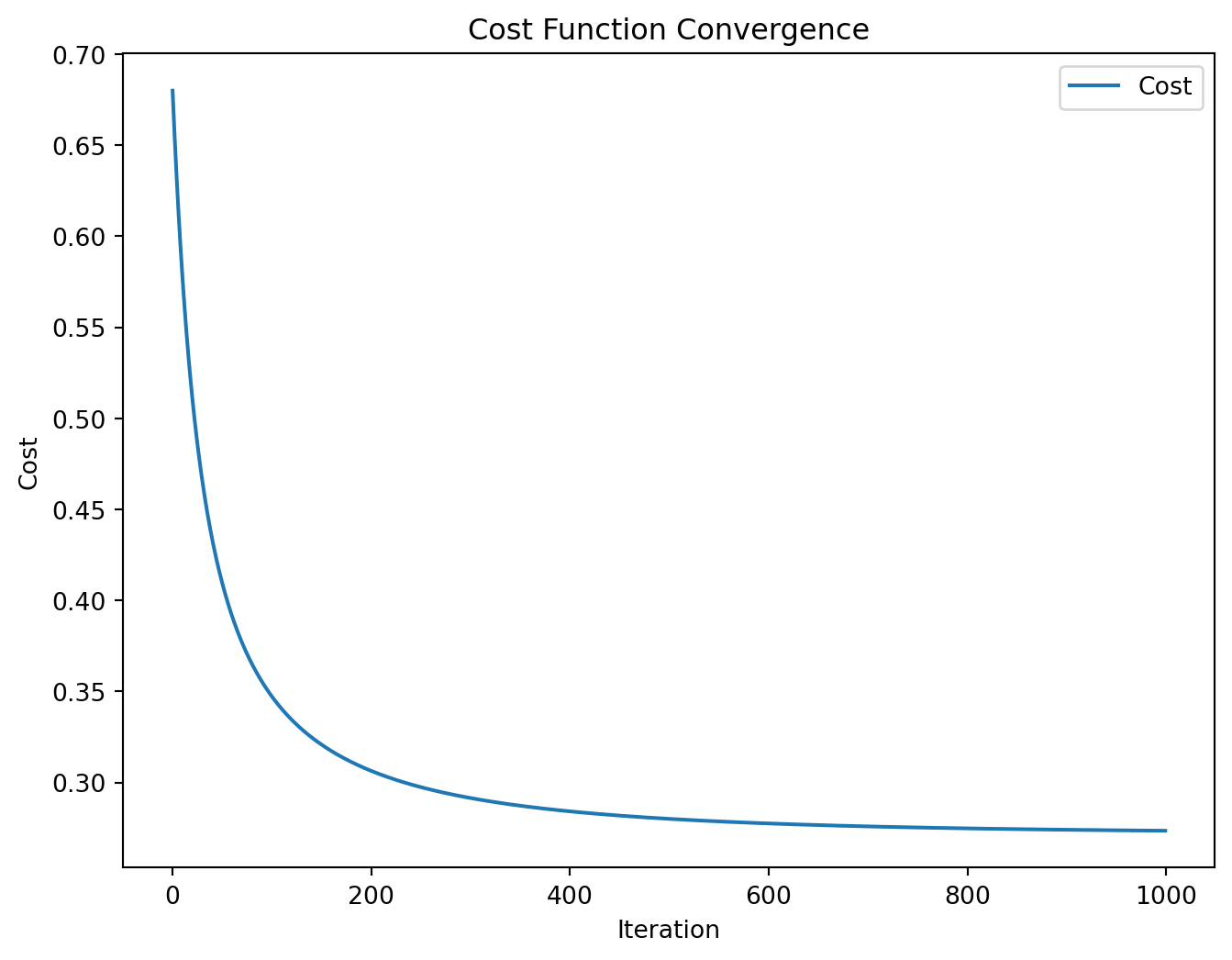
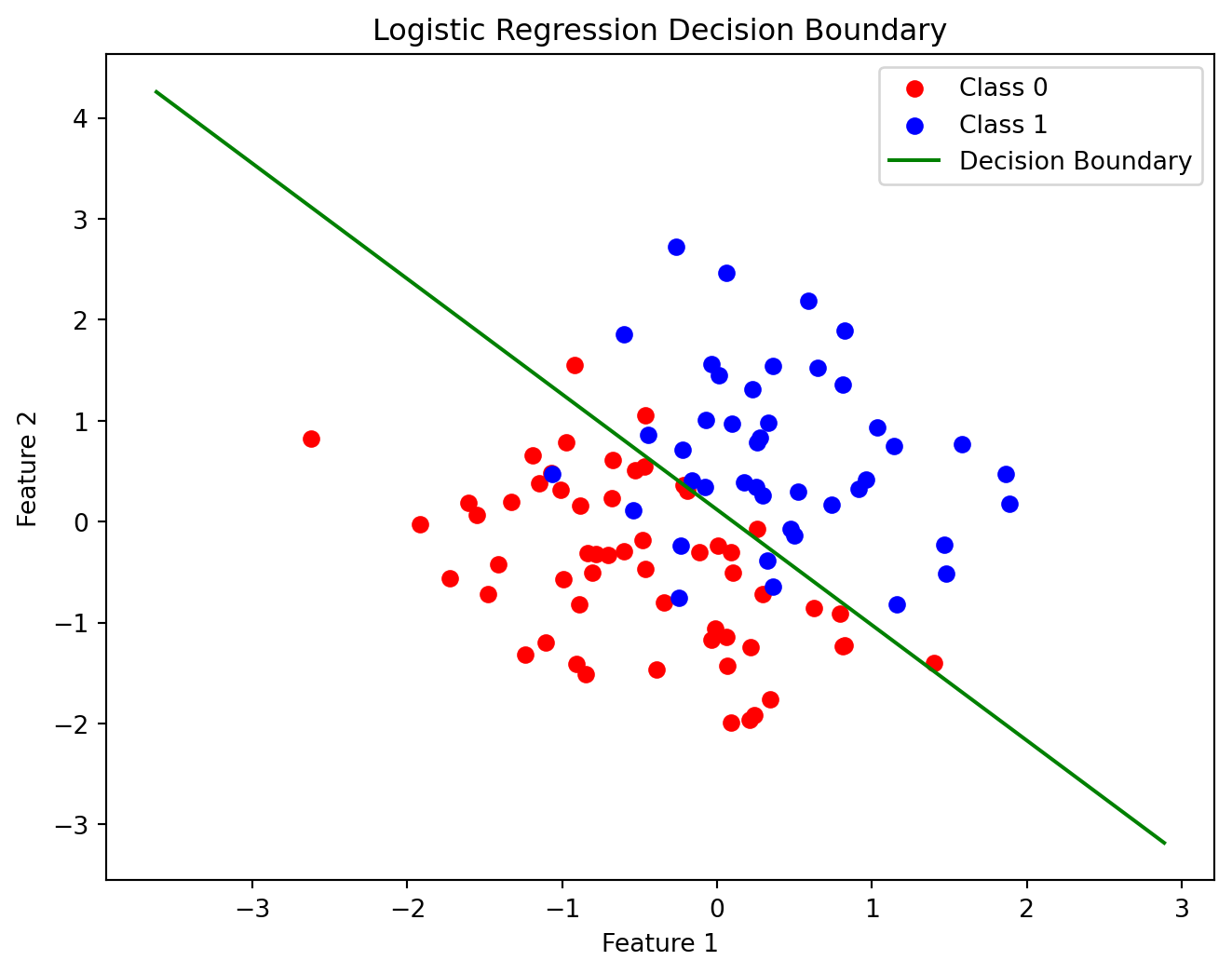
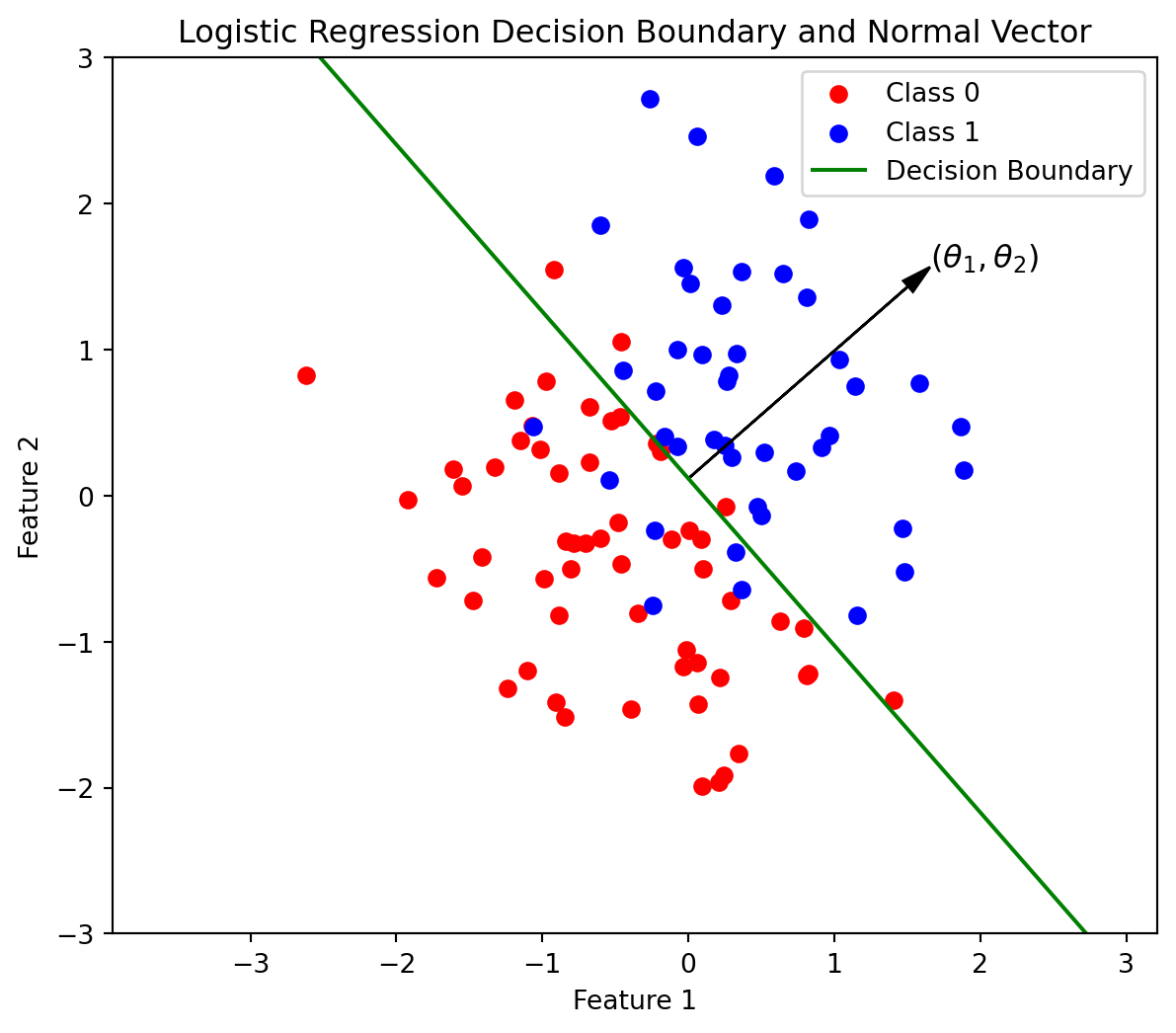
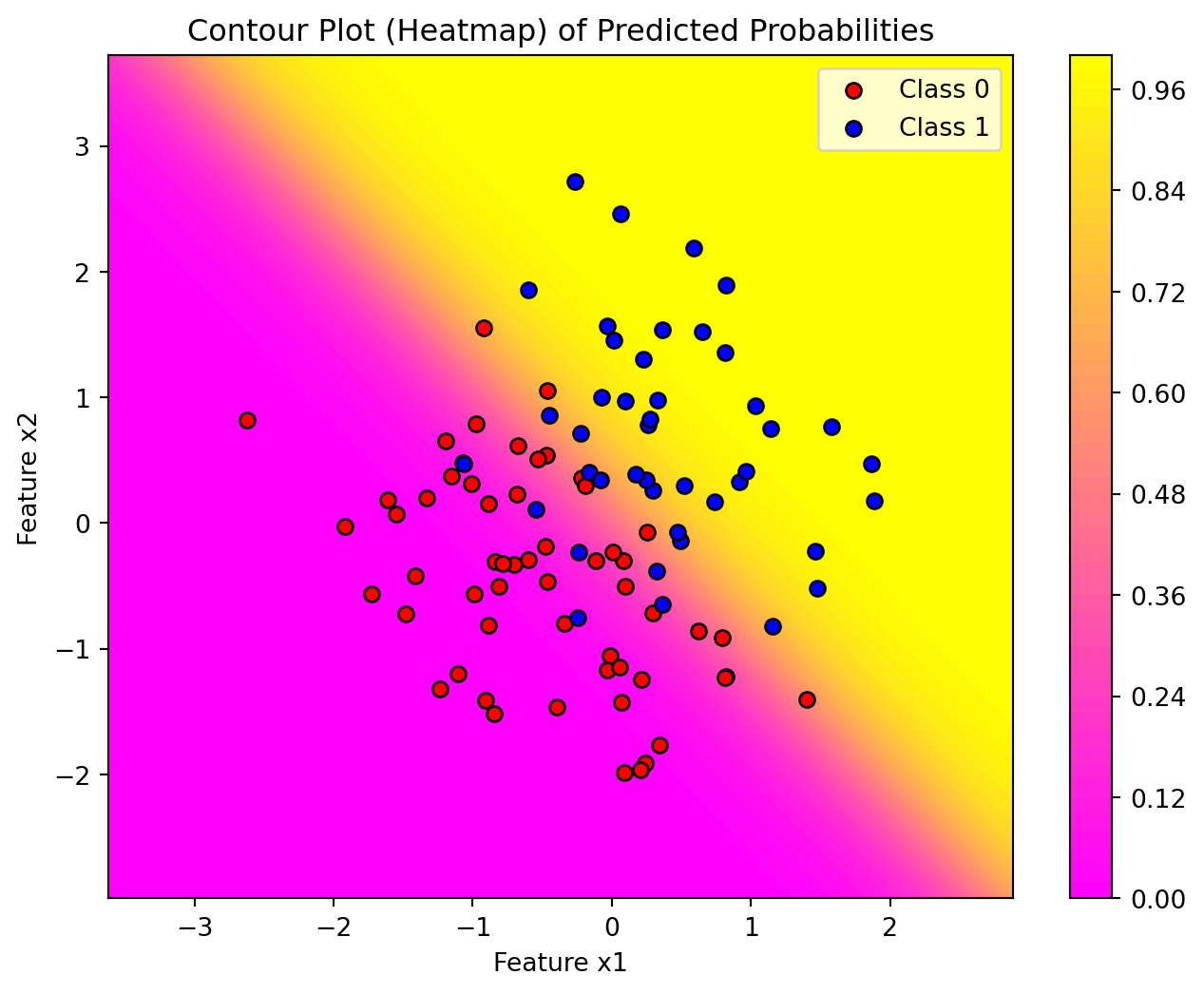
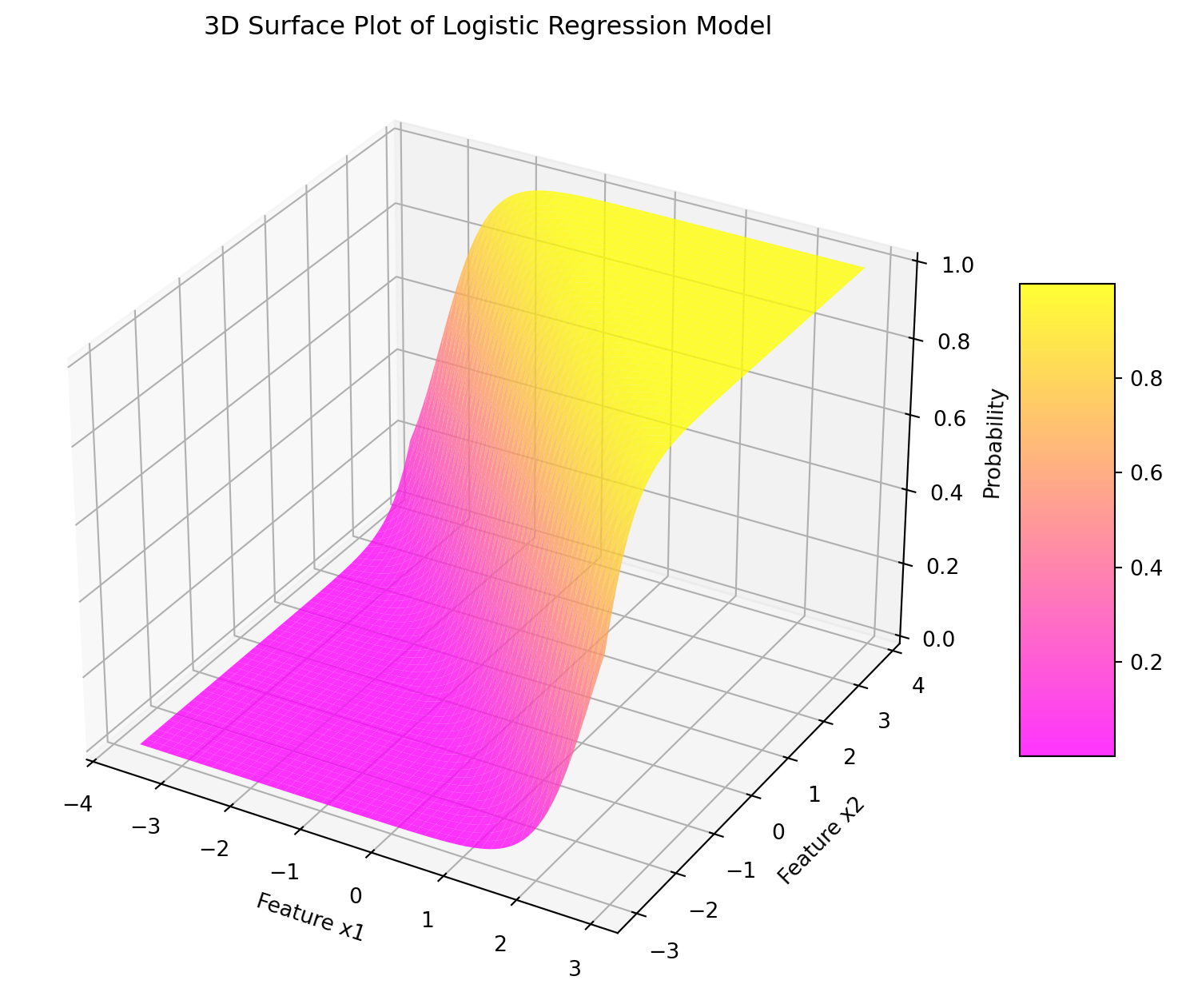
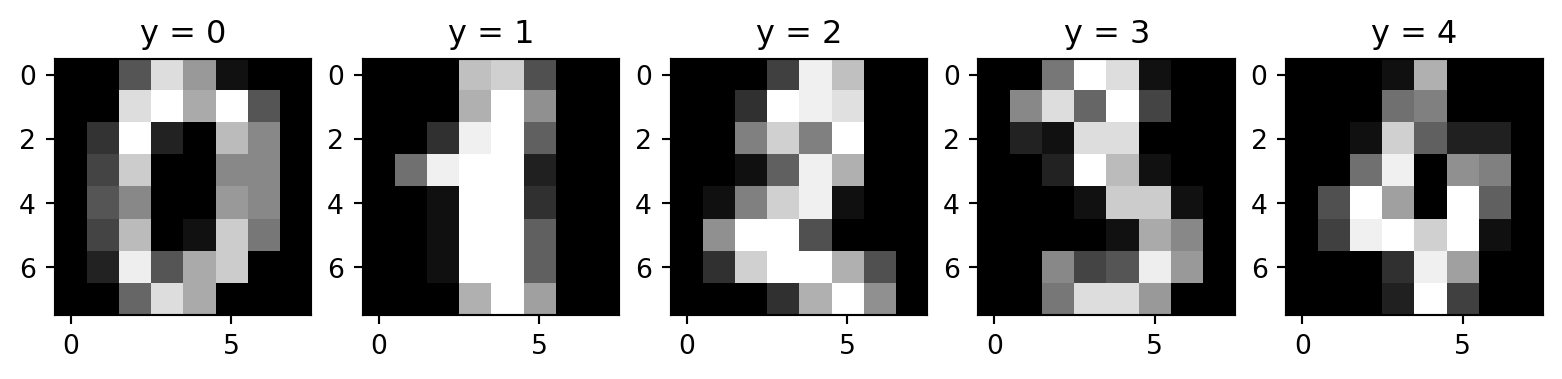
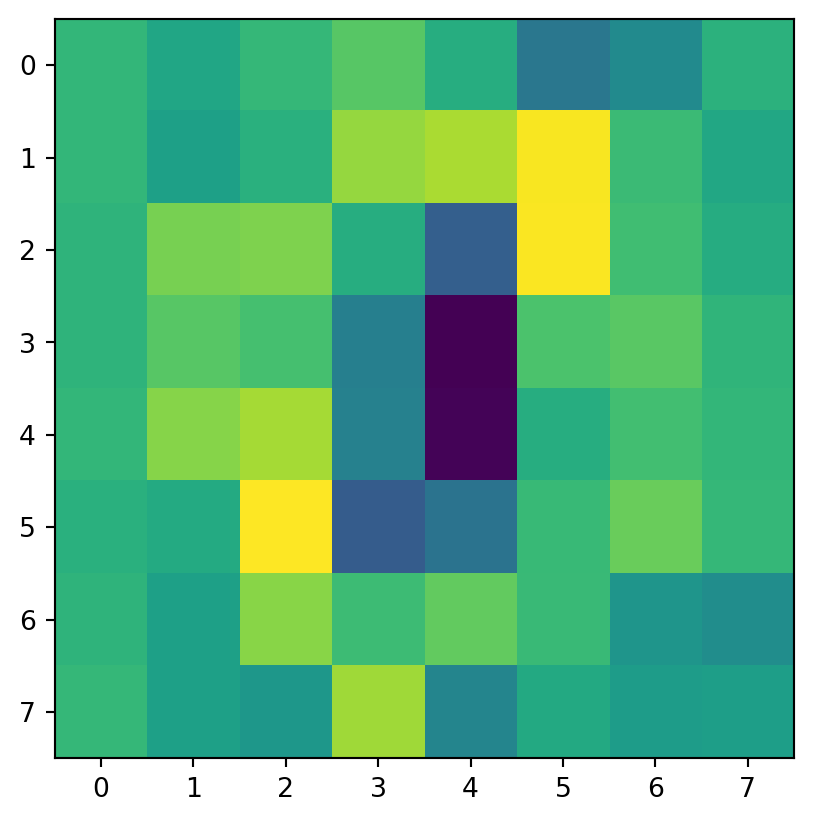
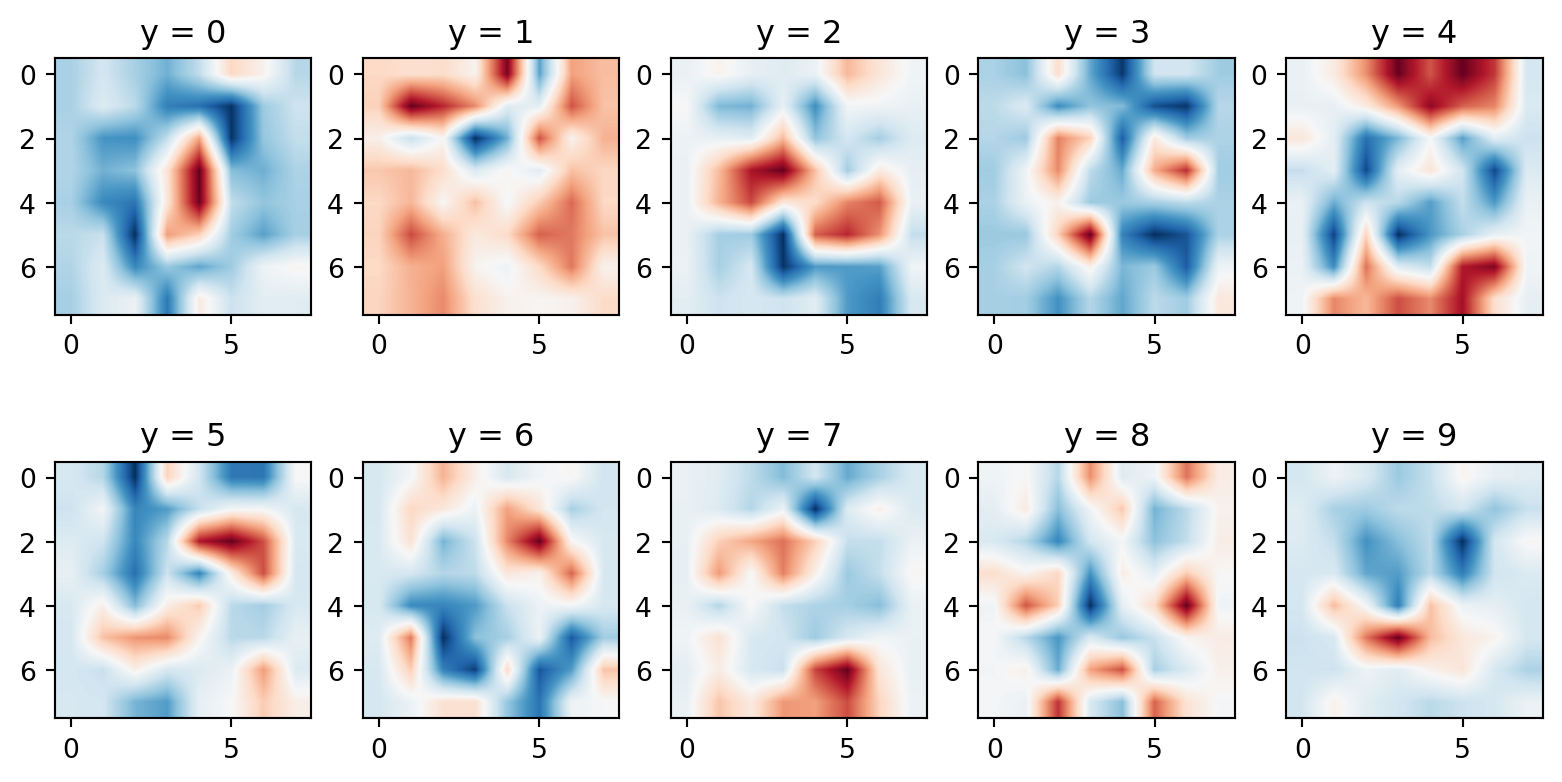
Code
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(42)
# Generate an array of p values from just above 0 to 1
p_values = np.linspace(0.001, 1, 1000)
# Compute the natural logarithm of each p value
ln_p_values = - np.log(p_values)
# Plot the graph
plt.figure(figsize=(5, 4))
plt.plot(p_values, ln_p_values, label=r'$-\log(\hat{y})$', color='b')
# Add labels and title
plt.xlabel(r'$\hat{y}$')
plt.ylabel(r'J')
plt.title(r'Graph of $-\log(\hat{y})$ for $\hat{y}$ from 0 to 1')
plt.grid(True)
plt.axhline(0, color='gray', lw=0.5) # Add horizontal line at y=0
plt.axvline(0, color='gray', lw=0.5) # Add vertical line at x=0
# Display the plot
plt.legend()
plt.show()