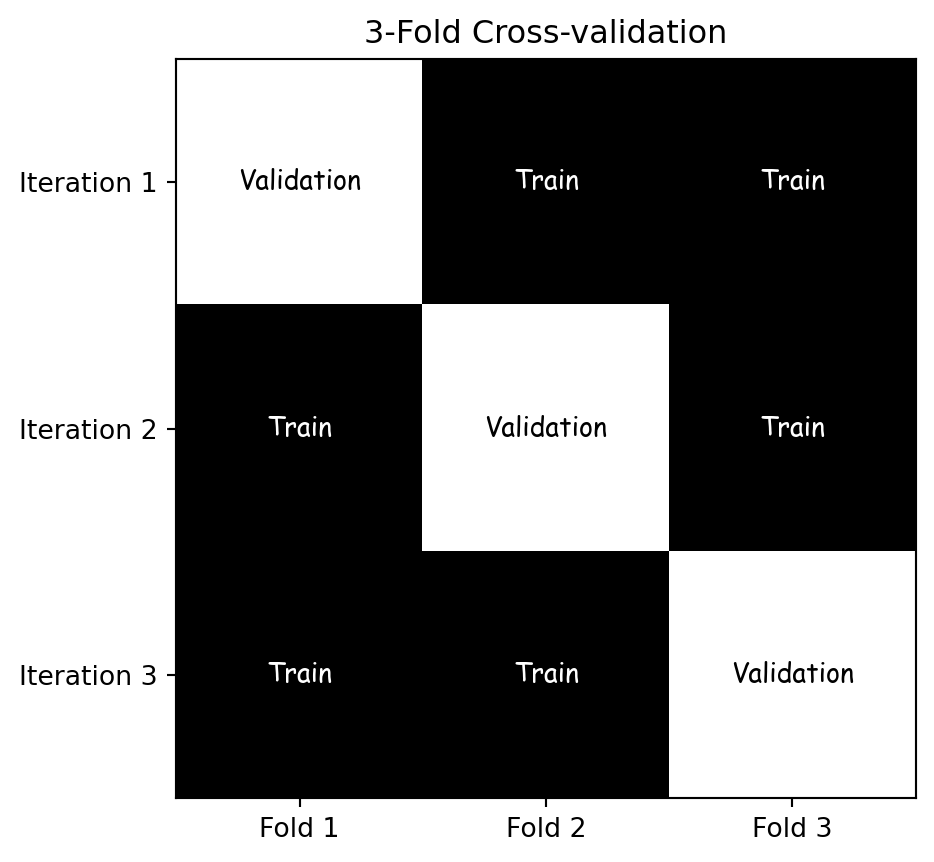
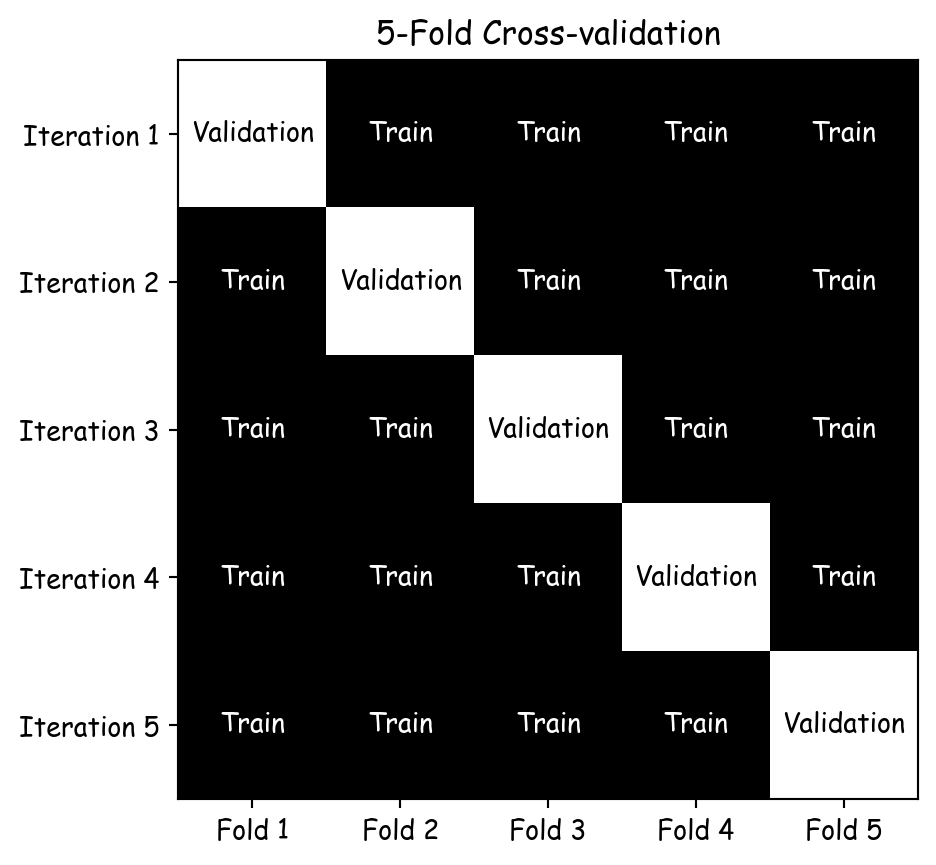
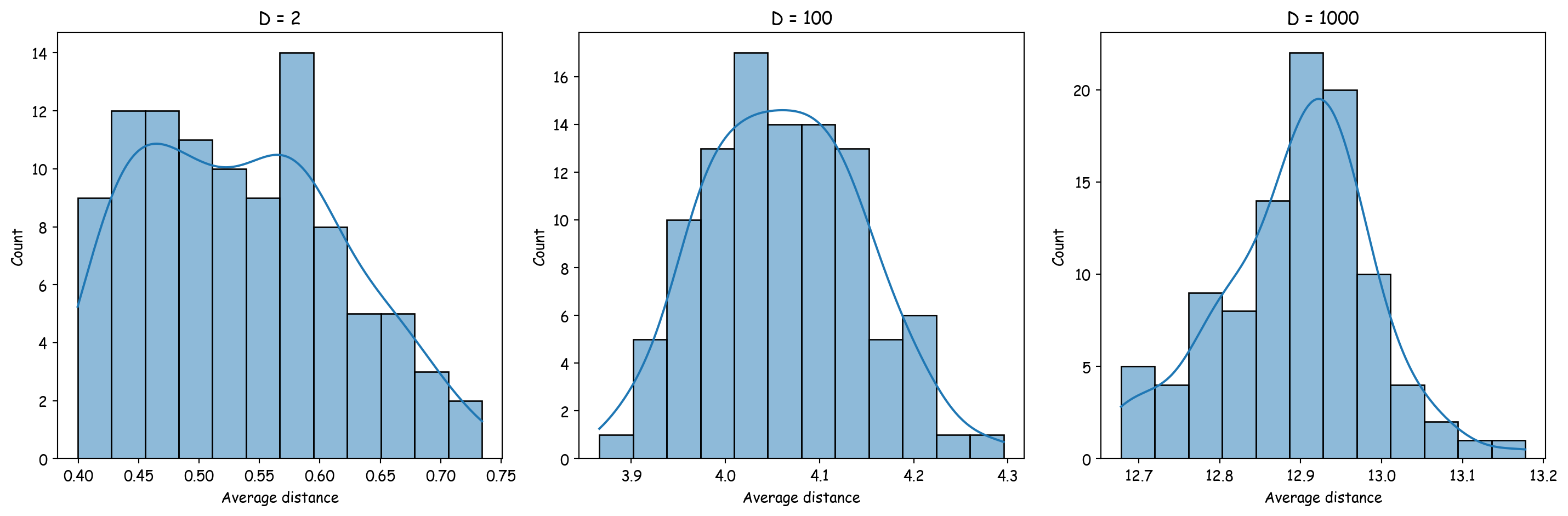
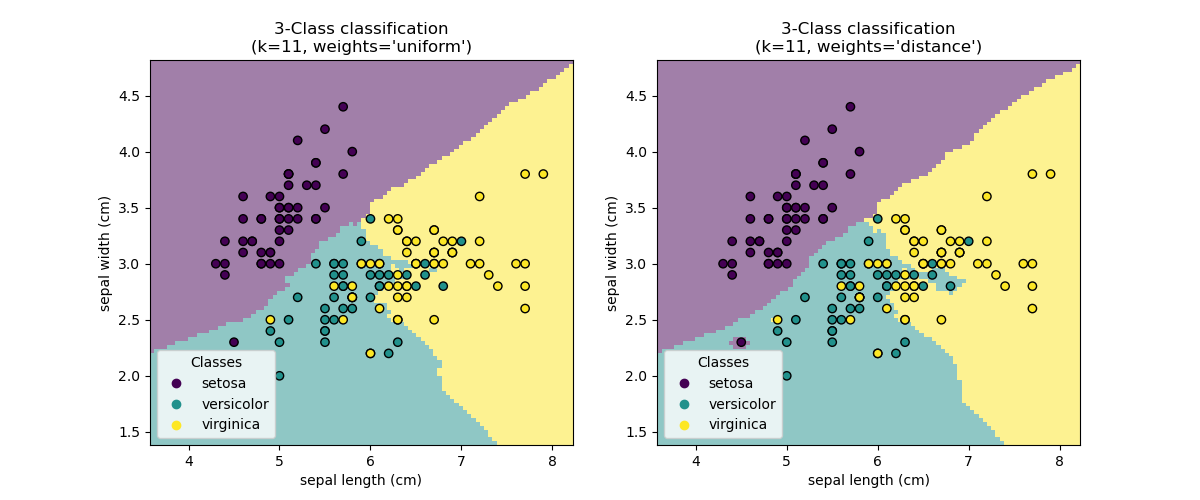
Altman, Naomi, and Martin Krzywinski. 2018.
“The curse(s) of dimensionality.” Nature Methods 15 (6): 399–400.
https://doi.org/10.1038/s41592-018-0019-x.
Japkowicz, Nathalie, and Mohak Shah. 2011. Evaluating Learning Algorithms: A Classification Perspective. Cambridge: Cambridge University Press.
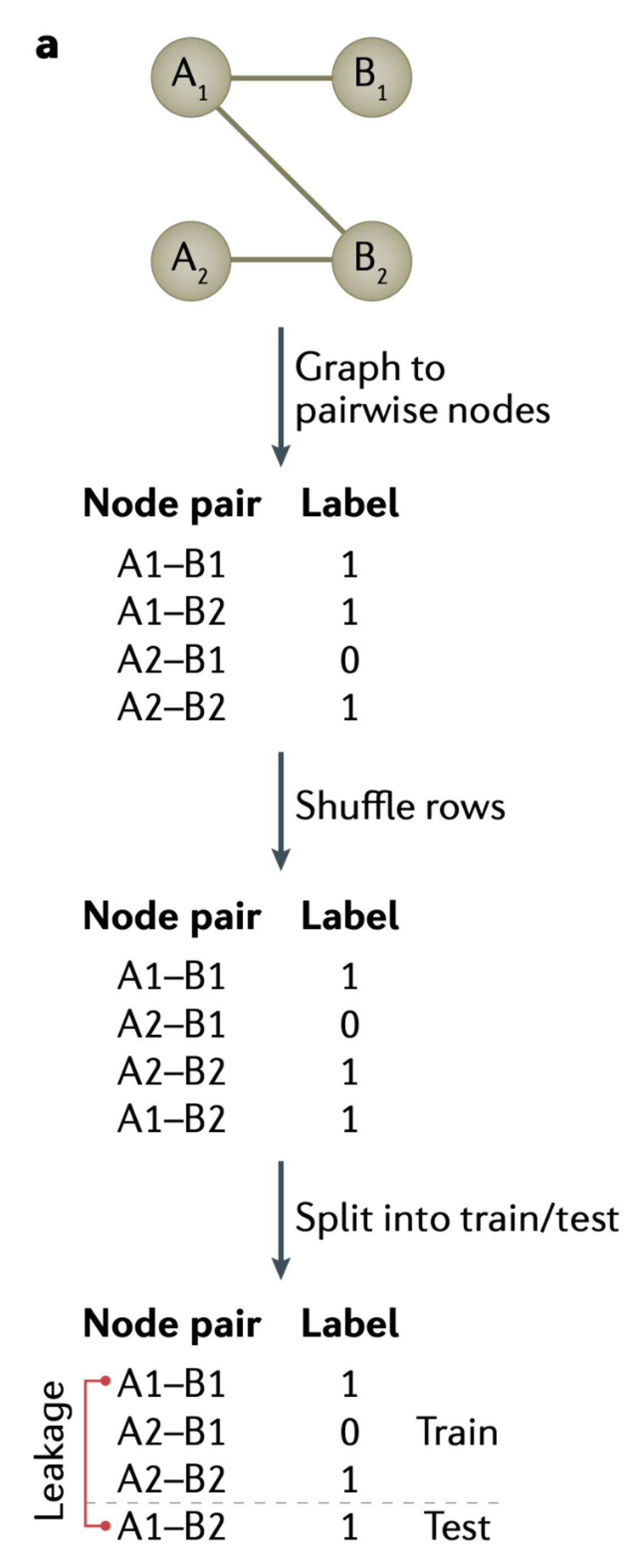
Rafi, Abdul Muntakim, Brett Kiyota, Nozomu Yachie, and Carl G de Boer. 2025.
“Detecting and avoiding homology-based data leakage in genome-trained sequence models.” https://doi.org/10.1101/2025.01.22.634321.
Sokolova, Marina, and Guy Lapalme. 2009.
“A systematic analysis of performance measures for classification tasks.” Information Processing and Management 45 (4): 427–37.
https://doi.org/10.1016/j.ipm.2009.03.002.
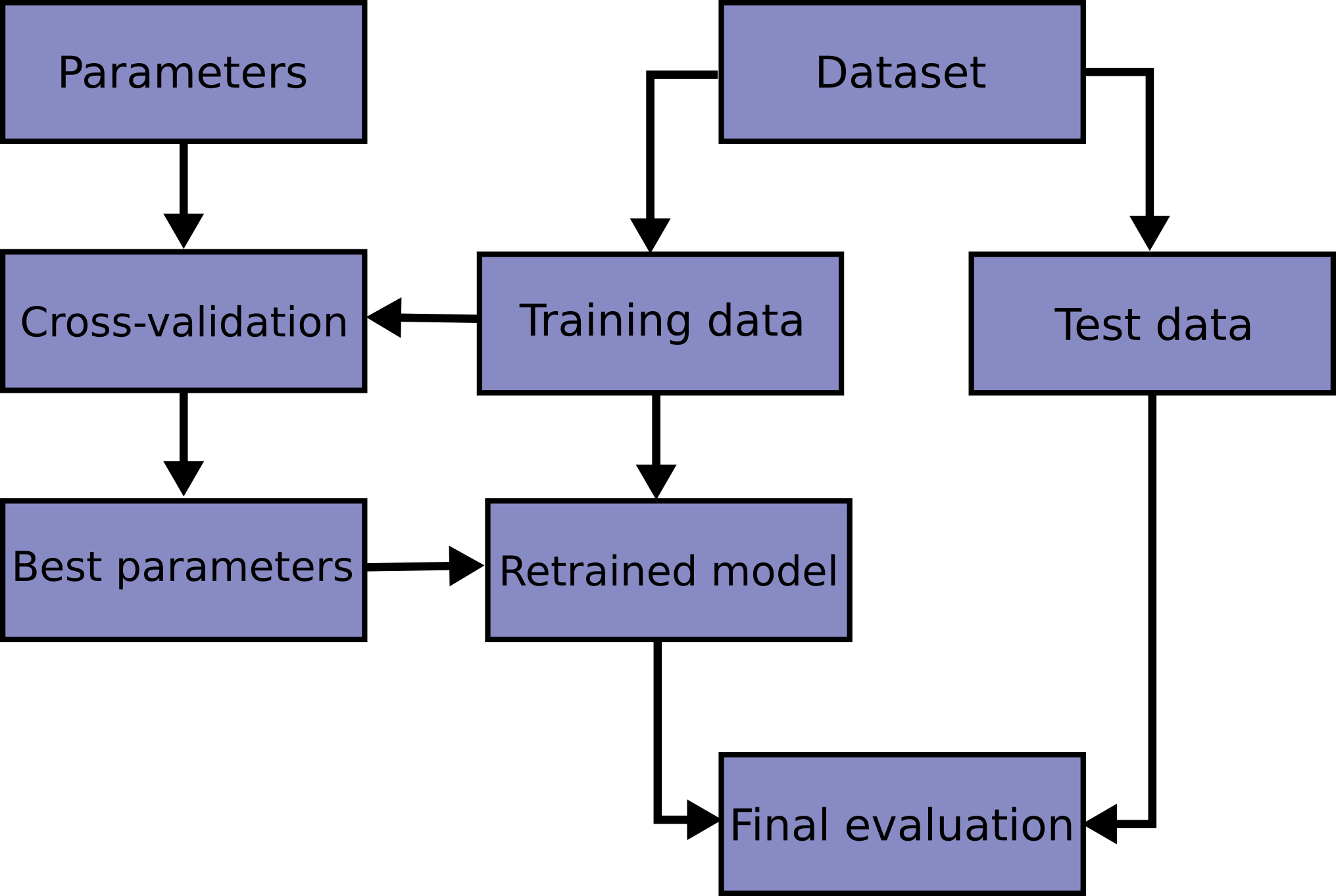
Walsh, Ian, Dmytro Fishman, Dario Garcia-Gasulla, Tiina Titma, Gianluca Pollastri, ELIXIR Machine Learning Focus Group, Emidio Capriotti, et al. 2021.
“DOME: recommendations for supervised machine learning validation in biology.” Nature Methods 18 (10): 1122–27.
https://doi.org/10.1038/s41592-021-01205-4.
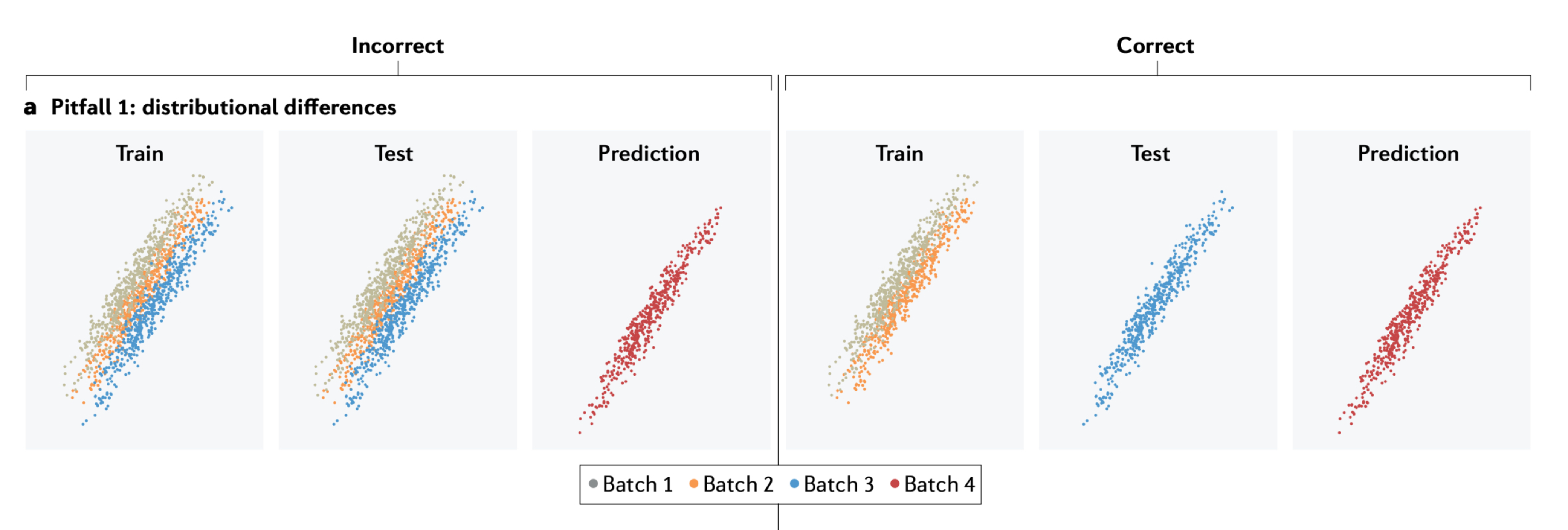
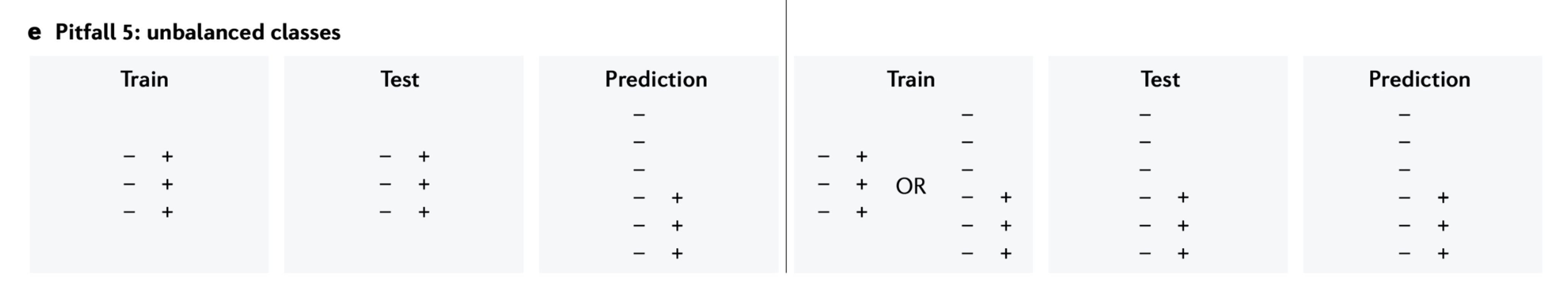
Whalen, Sean, Jacob Schreiber, William S. Noble, and Katherine S. Pollard. 2022.
“Navigating the pitfalls of applying machine learning in genomics.” Nature Reviews Genetics 23 (3): 169–81.
https://doi.org/10.1038/s41576-021-00434-9.