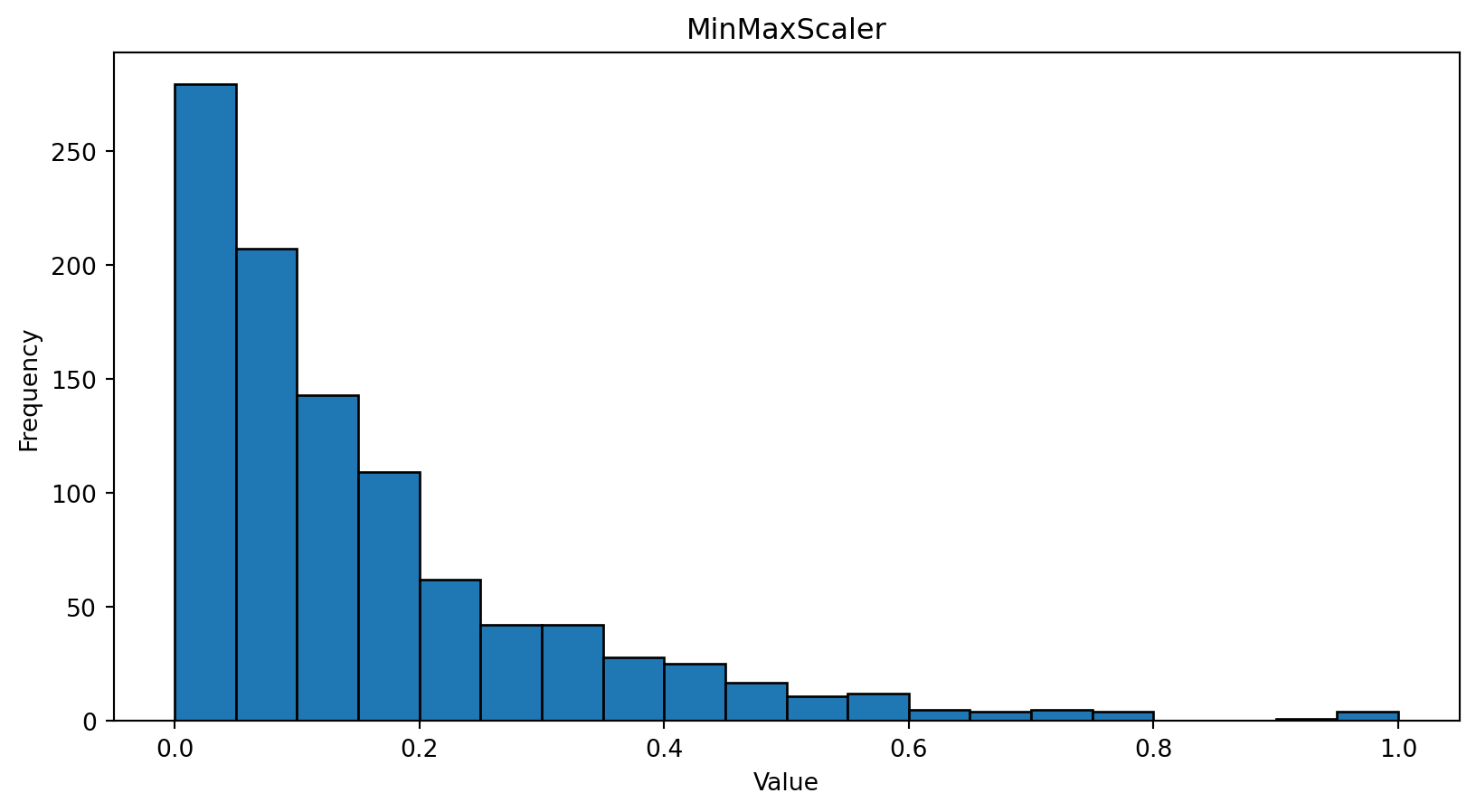
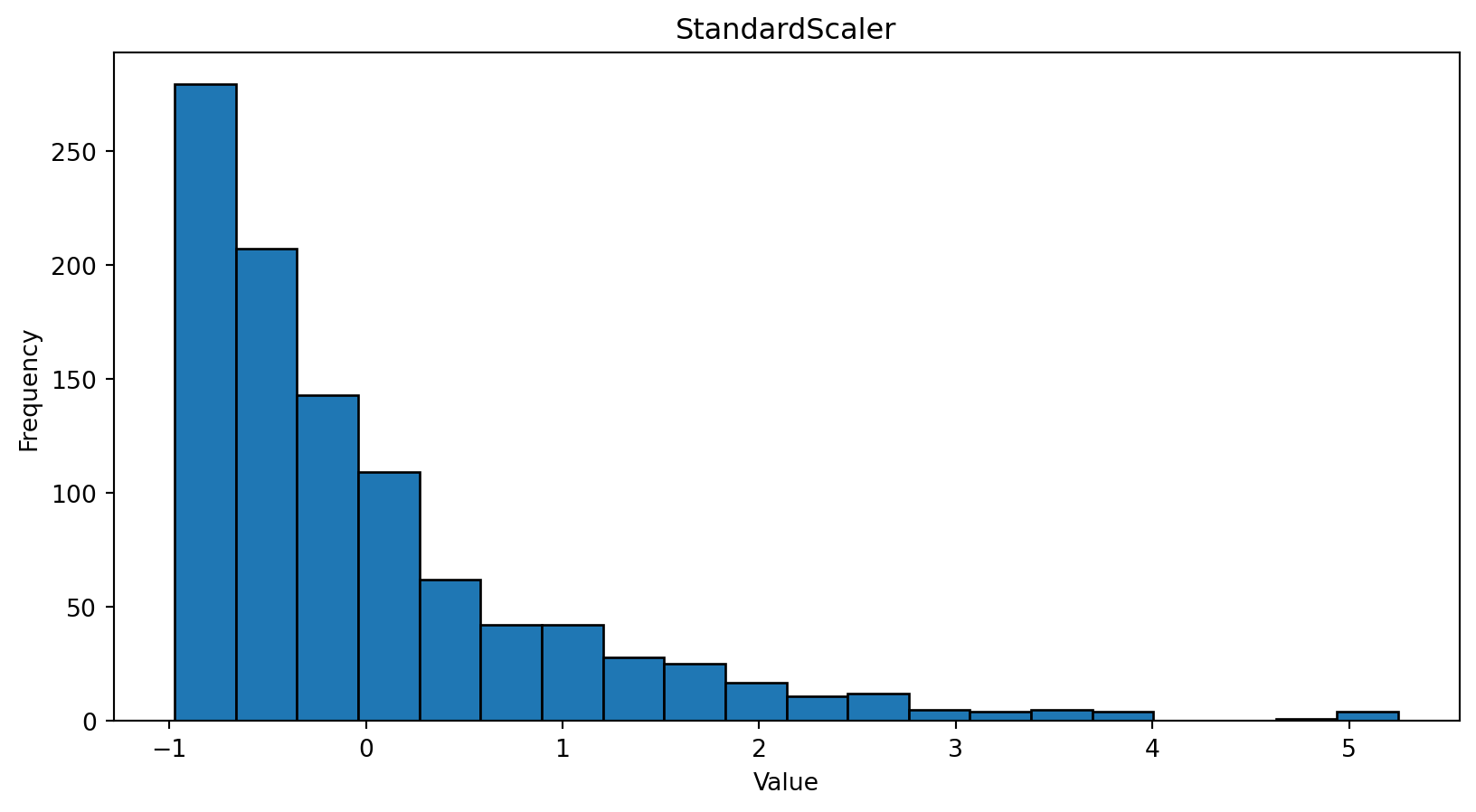
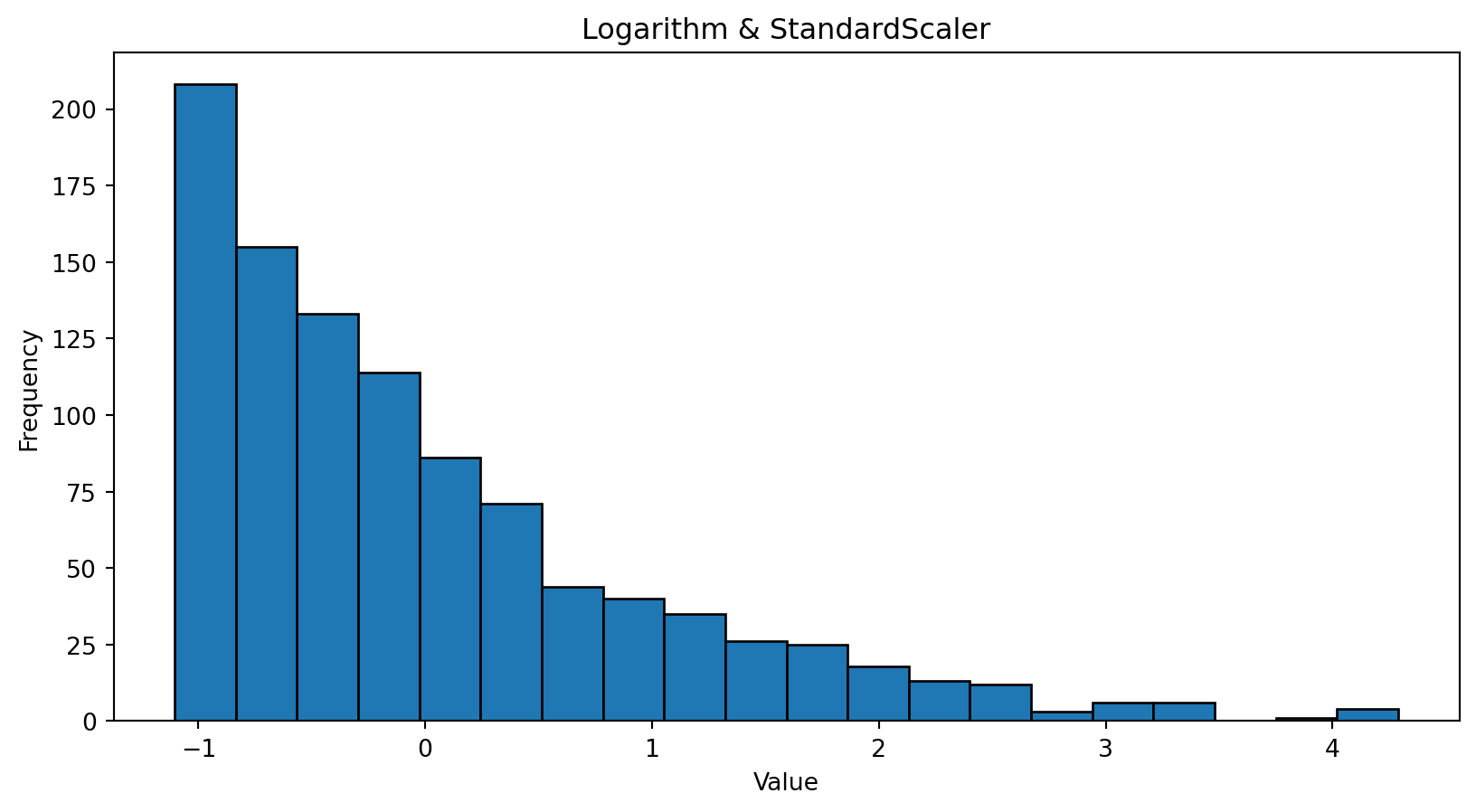
import numpy as np
np.random.seed(42)
from sklearn.datasets import fetch_openml
adult = fetch_openml(name='adult', version=2)
print(adult.DESCR)Author: Ronny Kohavi and Barry Becker
Source: UCI - 1996
Please cite: Ron Kohavi, “Scaling Up the Accuracy of Naive-Bayes Classifiers: a Decision-Tree Hybrid”, Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, 1996
Prediction task is to determine whether a person makes over 50K a year. Extraction was done by Barry Becker from the 1994 Census database. A set of reasonably clean records was extracted using the following conditions: ((AAGE>16) && (AGI>100) && (AFNLWGT>1)&& (HRSWK>0))
This is the original version from the UCI repository, with training and test sets merged.
Variable description
Variables are all self-explanatory except fnlwgt. This is a proxy for the demographic background of the people: “People with similar demographic characteristics should have similar weights”. This similarity-statement is not transferable across the 51 different states.
Description from the donor of the database:
The weights on the CPS files are controlled to independent estimates of the civilian noninstitutional population of the US. These are prepared monthly for us by Population Division here at the Census Bureau. We use 3 sets of controls. These are: 1. A single cell estimate of the population 16+ for each state. 2. Controls for Hispanic Origin by age and sex. 3. Controls by Race, age and sex.
We use all three sets of controls in our weighting program and “rake” through them 6 times so that by the end we come back to all the controls we used. The term estimate refers to population totals derived from CPS by creating “weighted tallies” of any specified socio-economic characteristics of the population. People with similar demographic characteristics should have similar weights. There is one important caveat to remember about this statement. That is that since the CPS sample is actually a collection of 51 state samples, each with its own probability of selection, the statement only applies within state.
Relevant papers
Ronny Kohavi and Barry Becker. Data Mining and Visualization, Silicon Graphics.
e-mail: ronnyk ‘@’ live.com for questions.
Downloaded from openml.org.